8PJM
 
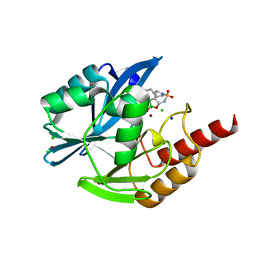 | |
7O6E
 
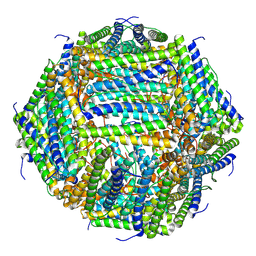 | | 2.12 A cryo-EM structure of Mycobacterium tuberculosis Ferritin | | Descriptor: | Ferritin BfrB | | Authors: | Gijsbers, A, Zhang, Y, Gao, Y, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-04-10 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Mycobacterium tuberculosis ferritin: a suitable workhorse protein for cryo-EM development.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7ZT6
 
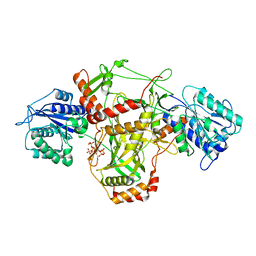 | |
3MU1
 
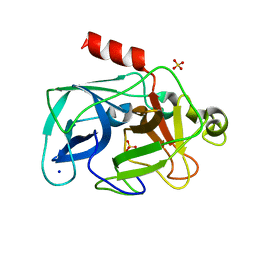 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6TOS
 
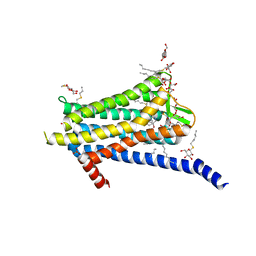 | | Crystal structure of the Orexin-1 receptor in complex with GSK1059865 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
4TT6
 
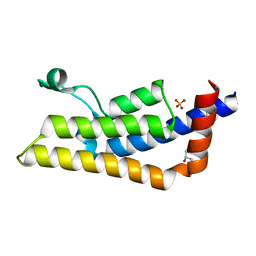 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
8CJO
 
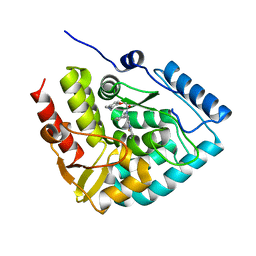 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-004 | | Descriptor: | 1-[(3~{S})-3-(4-chloranyl-2-fluoranyl-phenyl)-1,4,8-triazatricyclo[7.4.0.0^{2,7}]trideca-2(7),8-dien-4-yl]-2-(2-ethyl-6-methyl-pyridin-3-yl)oxy-ethanone, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86633706 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
6NHA
 
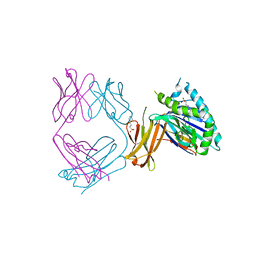 | | Crystal structure of SYNT001, a human FcRn blocking monoclonal antibody | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, IgG receptor FcRn large subunit p51, ... | | Authors: | Blumberg, R.S, Cheung, J, Mahmood, A, Gandhi, A.K. | | Deposit date: | 2018-12-21 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Blocking FcRn in humans reduces circulating IgG levels and inhibits IgG immune complex-mediated immune responses.
Sci Adv, 5, 2019
|
|
5HQD
 
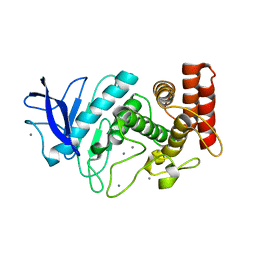 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Roesser, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
6NI4
 
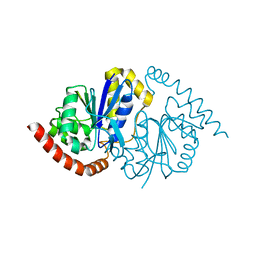 | |
5NPM
 
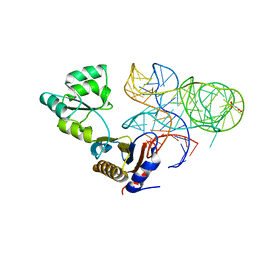 | | CRYSTAL STRUCTURE OF MUTANT RIBOSOMAL PROTEIN TTHL1 LACKING 8 N-TERMINAL RESIDUES IN COMPLEX WITH 80NT 23S RNA FROM THERMUS THERMOPHILUS | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, MALONATE ION, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Nevskaya, N.A, Nikonov, S.V, Garber, M.B. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | [Influence of Nonconserved Regions of L1 Protuberance of Thermus thermophilus Ribosome on the Affinity of L1 Protein to 23s rRNA].
Mol.Biol.(Moscow), 52, 2018
|
|
8BZZ
 
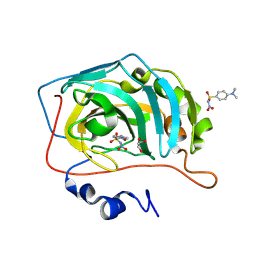 | |
4RKO
 
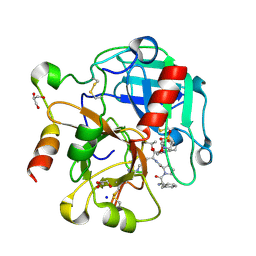 | | Crystal structure of thrombin mutant S195T bound with PPACK | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
8C3Z
 
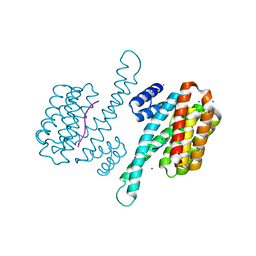 | |
7QY6
 
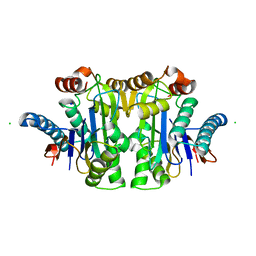 | | Structure of E.coli Class 2 L-asparaginase EcAIII, wild type (WT EcAIII) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1C4D
 
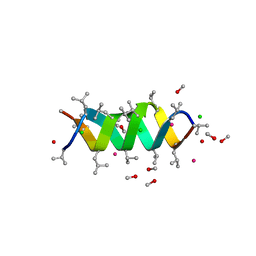 | | GRAMICIDIN CSCL COMPLEX | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Wallace, B.A. | | Deposit date: | 1999-06-04 | | Release date: | 2000-01-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Gramicidin Pore: Crystal Structure of a Cesium Complex.
Science, 241, 1988
|
|
4RMH
 
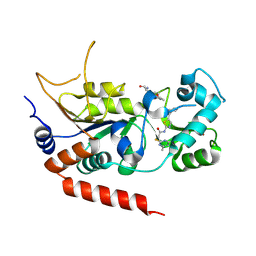 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
7NS9
 
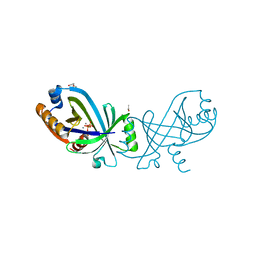 | | Triphosphate tunnel metalloenzyme from Sulfolobus acidocaldarius in complex with triphosphate and calcium | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vogt, M.S, Essen, L.-O, Banerjee, A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The archaeal triphosphate tunnel metalloenzyme SaTTM defines structural determinants for the diverse activities in the CYTH protein family.
J.Biol.Chem., 297, 2021
|
|
6Z2P
 
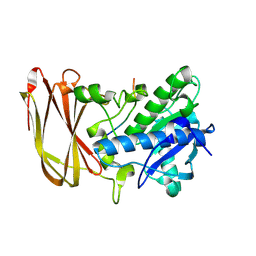 | | Crystal structure of catalytic inactive OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (glycodrosocin) substrate | | Descriptor: | CALCIUM ION, Glycodrosocin, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6QQD
 
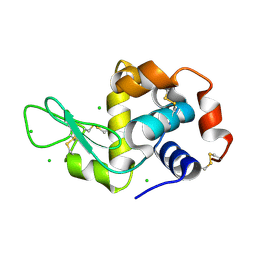 | |
7ZVZ
 
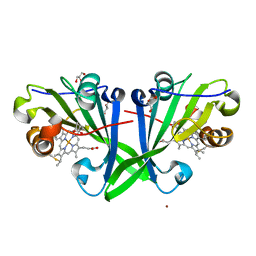 | |
6CCM
 
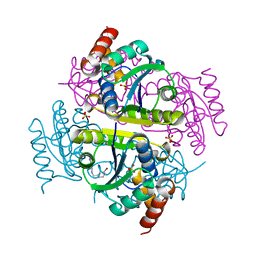 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-((3-bromobenzyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6R5F
 
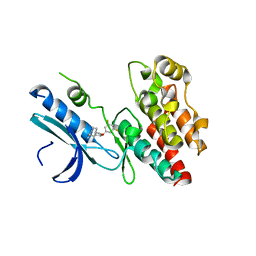 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
7ZVT
 
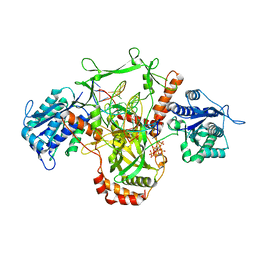 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
6R65
 
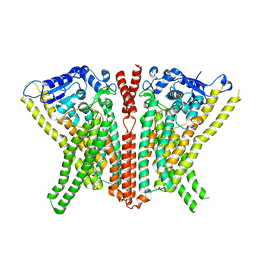 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
