6NVP
 
 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
7PC9
 
 | | The PDZ domain of SYNJ2BP complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, Protein Tax-1, Synaptojanin-2-binding protein,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6PU3
 
 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori | | Descriptor: | Heme-binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
6NLH
 
 | | Structure of human triose phosphate isomerase R189A | | Descriptor: | BROMIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Richards, K.R, Roland, B.P, Palladino, M.J, VanDemark, A.P. | | Deposit date: | 2019-01-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Missense variant in TPI1 (Arg189Gln) causes neurologic deficits through structural changes in the triosephosphate isomerase catalytic site and reduced enzyme levels in vivo.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
6NPZ
 
 | | Crystal structure of Akt1 (aa 123-480) kinase with a bisubstrate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | Authors: | Chu, N, Cole, P.A, Gabelli, S.B. | | Deposit date: | 2019-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
8FAR
 
 | |
6NW4
 
 | |
6QA2
 
 | | R80A MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Dautant, A, Henri, J, Wales, T.E, Meyer, P, Engen, J.R, Georgescauld, F. | | Deposit date: | 2018-12-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Remodeling of the Binding Site of Nucleoside Diphosphate Kinase Revealed by X-ray Structure and H/D Exchange.
Biochemistry, 58, 2019
|
|
6Q04
 
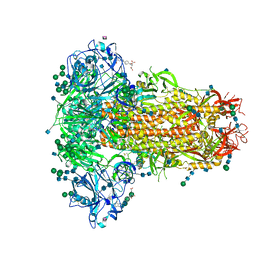 | | MERS-CoV S structure in complex with 5-N-acetyl neuraminic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NXJ
 
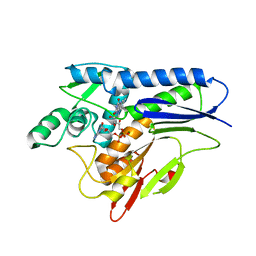 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6NY0
 
 | |
7MK0
 
 | |
7B8E
 
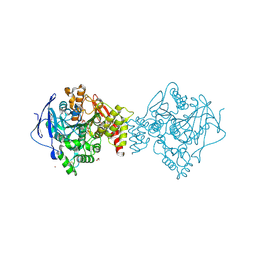 | | Torpedo californica acetylcholinesterase complexed with Ca+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L. | | Deposit date: | 2020-12-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
6QF1
 
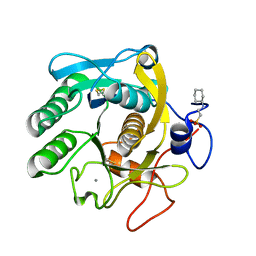 | | X-Ray structure of Proteinase K crystallized on a silicon chip | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Proteinase K | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6PV6
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2019-07-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
6PXC
 
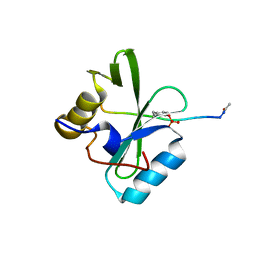 | |
6QAJ
 
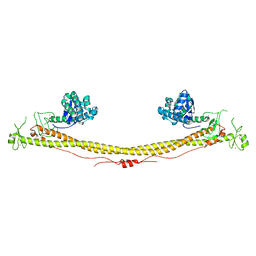 | | Structure of the tripartite motif of KAP1/TRIM28 | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Stoll, G.A, Oda, S, Yu, M, Modis, Y. | | Deposit date: | 2018-12-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7Q6N
 
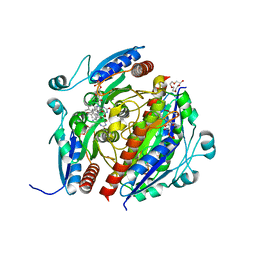 | | Structure of WrbA from Salmonella Typhimurium bound to ME0052 | | Descriptor: | 2-azanyl-4,6-bis(bromanyl)phenol, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone), ... | | Authors: | Gabrielsen, M, Beckham, K.S.H, Roe, A.J. | | Deposit date: | 2021-11-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds.
Microbiology (Reading, Engl.), 168, 2022
|
|
6O2Z
 
 | | Crystal structure of IDH1 R132H mutant in complex with compound 32 | | Descriptor: | 6-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methylpyridine-3-carbonitrile, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
7QY5
 
 | | Crystal structure of the S.pombe Ars2-Red1 complex. | | Descriptor: | NURS complex subunit pir2, RNA elimination defective protein Red1, ZINC ION | | Authors: | Foucher, A.E, Kadlec, J. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
6O3G
 
 | |
7MEP
 
 | |
8FQS
 
 | | ADC-212 in complex with boronic acid transition state inhibitor MB076 | | Descriptor: | 1-[(2R)-2-{2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetamido}-2-boronoethyl]-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Powers, R.A, Wallar, B.J, June, C.M. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
7QVH
 
 | |
6NBT
 
 | |
