4RT6
 
 | | Structure of a complex between hemopexin and hemopexin binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heme/hemopexin-binding protein, Hemopexin | | Authors: | Zambolin, S, Clantin, B, Haouz, A, Villeret, V, Delepelaire, P. | | Deposit date: | 2014-11-13 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for haem piracy from host haemopexin by Haemophilus influenzae.
Nat Commun, 7, 2016
|
|
7OR6
 
 | |
4RV5
 
 | | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with pyruvic acid | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with pyruvic acid
To be Published
|
|
7ORD
 
 | |
7P4I
 
 | |
6WSM
 
 | | Crystal structure of coiled coil region of human septin 8 | | Descriptor: | SULFATE ION, Septin-8 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
6WQU
 
 | | CSL (RBPJ) bound to Notch3 RAM and DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Neurogenic locus notch homolog protein 3, ... | | Authors: | Kovall, R.A, Gagliani, E, Hall, D. | | Deposit date: | 2020-04-29 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PIM-induced phosphorylation of Notch3 promotes breast cancer tumorigenicity in a CSL-independent fashion.
J.Biol.Chem., 296, 2021
|
|
4RK2
 
 | | Crystal structure of sugar transporter RHE_PF00321 from Rhizobium etli, target EFI-510806, an open conformation | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative sugar ABC transporter, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sugar Transporter RHE_Pf00321 from Rhizobium Etli, Target EFI-510806
To be Published
|
|
4RKR
 
 | | Crystal structure of LacI family transcriptional regulator from Arthrobacter sp. FB24, target EFI-560007, complex with lactose | | Descriptor: | Transcriptional regulator, LacI family, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator PurR from Arthrobacter Sp, Target Nysgxrc 11027R
To be Published
|
|
7P1E
 
 | | Structure of KDNase from Aspergillus Terrerus in complex with 2,3-difluoro-2-keto-3-deoxynononic acid | | Descriptor: | (2R,3R,4R,5R,6S)-2,3-bis(fluoranyl)-4,5-bis(oxidanyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
6YWV
 
 | | The structure of the Atp25 bound assembly intermediate of the mitoribosome from Neurospora crassa | | Descriptor: | 23 S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
7P1B
 
 | |
6WNM
 
 | | The structure of Pf4r from a superinfective isolate of the filamentous phage Pf4 of Pseudomonas aeruginosa PA01 | | Descriptor: | Pf4r, [(2R)-3-{[2-(carboxymethoxy)benzene-1-carbonyl]amino}-2-methoxypropyl](hydroxy)mercury | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
6WHN
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
7P75
 
 | |
4RVX
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL079 | | Descriptor: | (4S)-4-amino-N-[(2S,3S)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-3,4-dihydro-2H-chromene-6-carboxamide, HIV-1 Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2014-11-28 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1.
To be Published
|
|
4RWE
 
 | | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92 | | Descriptor: | CHLORIDE ION, GLYCEROL, Sugar-binding transport protein | | Authors: | Tan, K, Zhou, M, Clancy, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-03 | | Release date: | 2014-12-31 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92
To be Published
|
|
7OSE
 
 | | cytochrome bd-II type oxidase with bound aurachin D | | Descriptor: | Aurachin D, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-II ubiquinol oxidase subunit 1, ... | | Authors: | Grauel, A, Kaegi, J, Rasmussen, T, Wohlwend, D, Boettcher, B, Friedrich, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D.
Nat Commun, 12, 2021
|
|
4S0O
 
 | |
6WPN
 
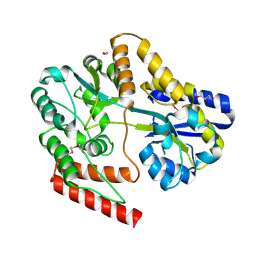 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Substrate-binding protein | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
4S0Z
 
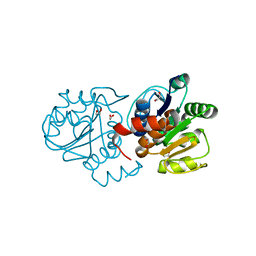 | | Crystal structure of M26V human DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Protein DJ-1 | | Authors: | Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2015-01-07 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transient sampling of aggregation-prone conformations causes pathogenic instability of a parkinsonian mutant of DJ-1 at physiological temperature.
Protein Sci., 24, 2015
|
|
7ONP
 
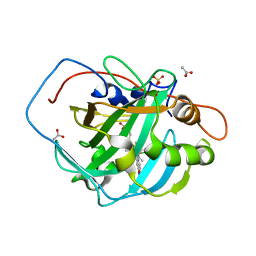 | | Wild type carbonic anhydrase II with bound IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-nitro-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
4RSM
 
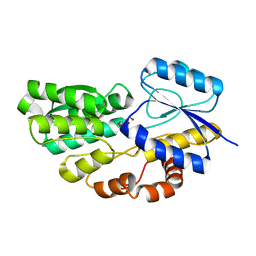 | | Crystal structure of carbohydrate transporter msmeg_3599 from mycobacterium smegmatis str. mc2 155, target efi-510970, in complex with d-threitol | | Descriptor: | D-Threitol, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4TQA
 
 | | Crystal Structure of a GDP-bound G13D Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
7OXO
 
 | | human LonP1, R-state, incubated in AMPPCP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
