4TUF
 
 | |
2FSW
 
 | | Crystal Structure of the Putative Transcriptional Regualator, MarR family from Porphyromonas gingivalis W83 | | Descriptor: | PG_0823 protein | | Authors: | Kim, Y, Quartey, P, Buelt, J, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Transcriptional Regualator, MarR family from Porphyromonas gingivalis W83
To be Published
|
|
4TVG
 
 | | HIV Protease (PR) dimer in closed form with pepstatin in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, GLYCEROL, HIV Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
4TXD
 
 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
4TZA
 
 | |
4U06
 
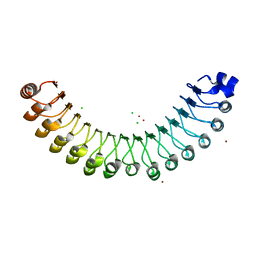 | | Structure of Leptospira interrogans LRR protein LIC10831 | | Descriptor: | CHLORIDE ION, LIC10831, ZINC ION | | Authors: | Shepard, W, Saul, F.A, Haouz, A, Picardeau, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a novel subfamily of leucine-rich repeat proteins from the human pathogen Leptospira interrogans.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U08
 
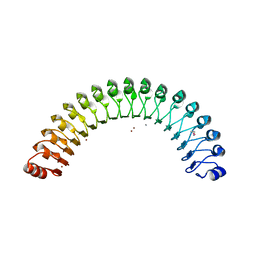 | | Structure of Leptospira interrogans LRR protein LIC11098 | | Descriptor: | CALCIUM ION, LIC11098, SULFATE ION, ... | | Authors: | Shepard, W, Saul, F.A, Haouz, A, Picardeau, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of a novel subfamily of leucine-rich repeat proteins from the human pathogen Leptospira interrogans.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U0B
 
 | |
4U0F
 
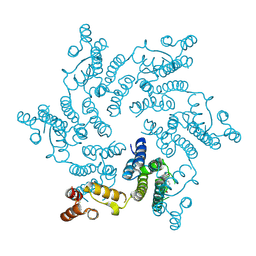 | | Hexameric HIV-1 CA in Complex with BI-2 | | Descriptor: | (4S)-4-(4-hydroxyphenyl)-3-phenyl-4,5-dihydropyrrolo[3,4-c]pyrazol-6(1H)-one, 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4TXO
 
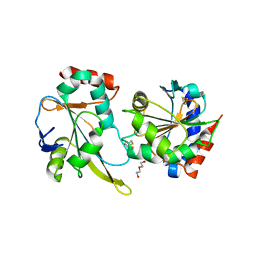 | | Crystal structure of the mixed disulfide complex of thioredoxin-like TlpAs(C110S) and copper chaperone ScoIs(C74S) | | Descriptor: | Blr1131 protein, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Scharer, M.A, Abicht, H.K, Glockshuber, R, Hennecke, H. | | Deposit date: | 2014-07-04 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Periplasmic Thioredoxin TlpA Reduces Bacterial Copper Chaperone ScoI and Cytochrome Oxidase Subunit II (CoxB) Prior to Metallation.
J.Biol.Chem., 289, 2014
|
|
4U4W
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4TZG
 
 | | Crystal structure of eCGP123, an extremely thermostable green fluorescent protein | | Descriptor: | Fluorescent Protein | | Authors: | Close, D.W, Don Paul, C, Traore, D.A.K, Wilce, M.C.J, Prescott, M, Bradbury, A.R.M. | | Deposit date: | 2014-07-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering.
Proteins, 83, 2015
|
|
4U0D
 
 | | Hexameric HIV-1 CA in complex with Nup153 peptide, P212121 crystal form | | Descriptor: | CHLORIDE ION, Gag polyprotein, Nuclear pore complex protein Nup153 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U9R
 
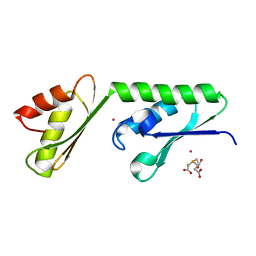 | | Structure of the N-terminal Extension from Cupriavidus metallidurans CzcP | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, CADMIUM ION, CzcP cation efflux P1-ATPase | | Authors: | Smith, A.T, Rosenzweig, A.C. | | Deposit date: | 2014-08-06 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A new metal binding domain involved in cadmium, cobalt and zinc transport.
Nat.Chem.Biol., 11, 2015
|
|
4U24
 
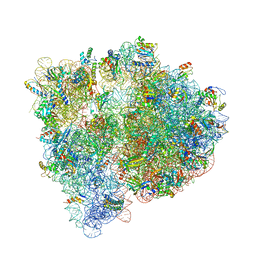 | | Crystal structure of the E. coli ribosome bound to dalfopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4TZV
 
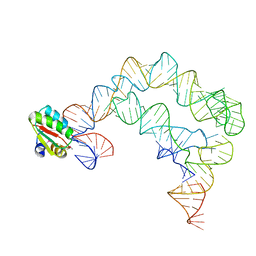 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate post crystallization | | Descriptor: | Ribosome-associated protein L7Ae-like, T-box stem I, engineered tRNA | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (5.03 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
7C3T
 
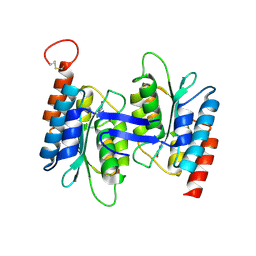 | | Crystal structure of NE0047 (N66Q) mutant in complex with 8-azaguanine | | Descriptor: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | Authors: | Gaded, V, Bitra, A, Singh, J, Anand, R. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure guided mutagenesis reveals the substrate determinants of guanine deaminase.
J.Struct.Biol., 213, 2021
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
7C3U
 
 | | Crystal structure of NE0047 (N66A) mutant in complex with 8-azaguanine | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Gaded, V, Bitra, A, Singh, J, Anand, R. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure guided mutagenesis reveals the substrate determinants of guanine deaminase.
J.Struct.Biol., 213, 2021
|
|
6KZM
 
 | | GluK3 receptor complex with kainate | | Descriptor: | Glutamate receptor ionotropic, kainate 3 | | Authors: | Kumar, J, Kumari, J, Burada, A.P. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural dynamics of the GluK3-kainate receptor neurotransmitter binding domains revealed by cryo-EM.
Int.J.Biol.Macromol., 149, 2020
|
|
4U5Q
 
 | | High resolution crystal structure of reductase (R) domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Peptide synthetase | | Authors: | Patel, K.D, Haque, A.S, Priyadarshan, K, Sankaranarayanan, R. | | Deposit date: | 2014-07-25 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | High resolution structure of R-domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis
To Be Published
|
|
4U6V
 
 | | Mechanisms of Neutralization of a Human Anti-Alpha Toxin Antibody | | Descriptor: | Alpha-hemolysin, Fab, antigen binding fragment, ... | | Authors: | Oganesyan, V.Y, Peng, L, Damschroder, M.M, Cheng, L, Sadowska, A, Tkaczyk, C, Sellman, B, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2014-07-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanisms of Neutralization of a Human Anti-alpha-toxin Antibody.
J.Biol.Chem., 289, 2014
|
|
6L6F
 
 | | GluK3 receptor complex with UBP301 | | Descriptor: | Glutamate receptor ionotropic, kainate 3 | | Authors: | Kumar, J, Kumari, J, Burada, A.P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structural dynamics of the GluK3-kainate receptor neurotransmitter binding domains revealed by cryo-EM.
Int.J.Biol.Macromol., 149, 2020
|
|
4U6H
 
 | | Vaccinia L1/M12B9-Fab complex | | Descriptor: | Heavy chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Light chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Protein L1 | | Authors: | Matho, M.H, Schlossman, A, Zajonc, D.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent neutralization of vaccinia virus by divergent murine antibodies targeting a common site of vulnerability in l1 protein.
J.Virol., 88, 2014
|
|
2GGQ
 
 | | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii | | Descriptor: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase, IODIDE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii
To be published
|
|
