6S74
 
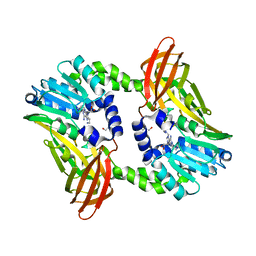 | | Crystal structure of CARM1 in complex with inhibitor UM305 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-azanylpropyl-[3-(pyrimidin-2-ylamino)propyl]amino]methyl]oxolane-3,4-diol, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S7E
 
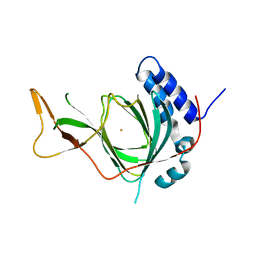 | | Plant Cysteine Oxidase PCO4 from Arabidopsis thaliana (using PEG 3350 and NaF as precipitants) | | Descriptor: | FE (III) ION, Plant cysteine oxidase 4 | | Authors: | White, M.D, Levy, C.W, Flashman, E, McDonough, M.A. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5QPI
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000554a | | Descriptor: | 5-methoxy-2-(1~{H}-pyrazol-3-yl)phenol, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
4O45
 
 | | WDR5 in complex with influenza NS1 C-terminal tail | | Descriptor: | Nonstructural protein 1, UNKNOWN ATOM OR ION, WD repeat-containing protein 5 | | Authors: | Qin, S, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
5NQG
 
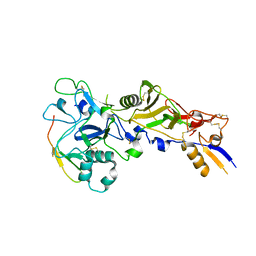 | | Crystal structure of Plasmodium vivax AMA1 in complex with a 39 aa PvRON2 peptide | | Descriptor: | Apical merozoite antigen 1, RON2 | | Authors: | Vulliez-le Normand, B, Saul, F.A, Faber, B.W, Bentley, G.A. | | Deposit date: | 2017-04-20 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cross-reactivity between apical membrane antgen 1 and rhoptry neck protein 2 in P. vivax and P. falciparum: A structural and binding study.
PLoS ONE, 12, 2017
|
|
2VEQ
 
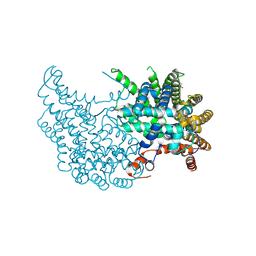 | |
6CJG
 
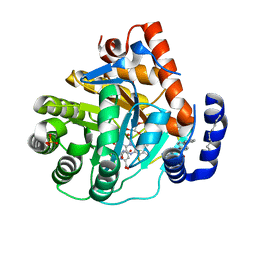 | | Human dihydroorotate dehydrogenase bound to napthyridine inhibitor 46 | | Descriptor: | 2-([1,1'-biphenyl]-4-yl)-3-methyl-1,7-naphthyridine-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
4ZNY
 
 | |
6MGY
 
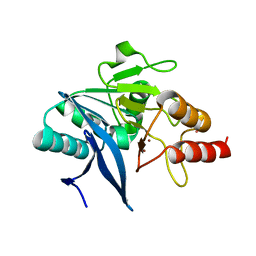 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae | | Descriptor: | GLYCEROL, Metallo-beta-lactamase NDM-5, POTASSIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae
To Be Published
|
|
2VAS
 
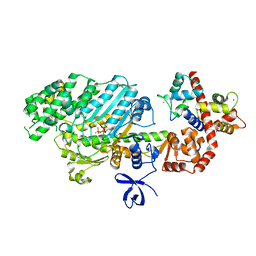 | | Myosin VI (MD-insert2-CaM, Delta-Insert1) Post-rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
6MHV
 
 | | Structure of human TRPV3 in the presence of 2-APB in C4 symmetry | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
5KBA
 
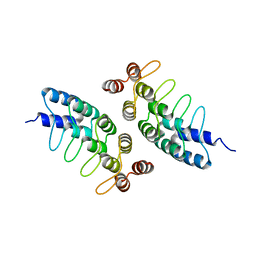 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein ANK1C2 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-06-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
8OMK
 
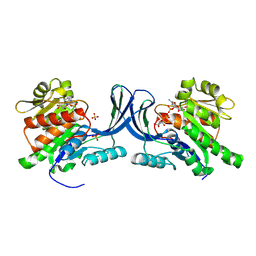 | | hKHK-C in complex with ADP & fructose 1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, ... | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
6FZ1
 
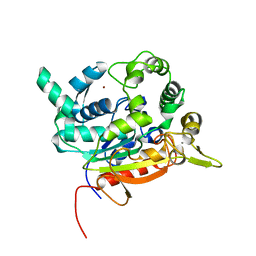 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6MK1
 
 | | Cryo-EM of self-assembly peptide filament HEAT_R1 | | Descriptor: | peptide HEAT_R1 | | Authors: | Wang, F, Hughes, S.A, Orlova, A, Conticello, V.P, Egelman, E.H. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Ambidextrous helical nanotubes from self-assembly of designed helical hairpin motifs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8VEM
 
 | | IsPETase - ACCE mutant | | Descriptor: | Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
5N2D
 
 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[2,6-dimethoxy-4-[(2-methyl-3-phenyl-phenyl)methoxy]phenyl]methylamino]ethyl]ethanamide | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
4Y44
 
 | | Endothiapepsin in complex with fragment 164 | | Descriptor: | 4,6-dimethyl-2-{[3-(morpholin-4-yl)propyl]amino}pyridine-3-carbonitrile, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
3S73
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S71
 
 | |
5N3K
 
 | |
5KEM
 
 | | EBOV sGP in complex with variable Fab domains of IgGs c13C6 and BDBV91 | | Descriptor: | BDBV91 variable Fab domain heavy chain, BDBV91 variable Fab domain light chain, Ebola secreted glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
6CMM
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor
To Be Published
|
|
6N32
 
 | | Anti-HIV-1 Fab 2G12 re-refinement | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, SULFATE ION | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
7YK4
 
 | | ox40-antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 4, antibody-H, ... | | Authors: | Zhou, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of a Novel Agonistic Anti-OX40 Antibody.
Biomolecules, 12, 2022
|
|
