6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
8QN6
 
 | | Amyloid-beta 40 type 2 filament from the leptomeninges of individual with Alzheimer's disease and cerebral amyloid angiopathy | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Yang, Y, Murzin, A.S, Peak-Chew, S.Y, Franco, C, Newell, K.L, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2023-09-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structures of A beta 40 filaments from the leptomeninges of individuals with Alzheimer's disease and cerebral amyloid angiopathy.
Acta Neuropathol Commun, 11, 2023
|
|
8TU9
 
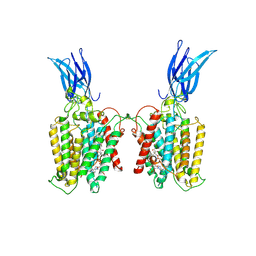 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N -acetyltransferase (HGSNAT).
Elife, 13, 2024
|
|
8QCL
 
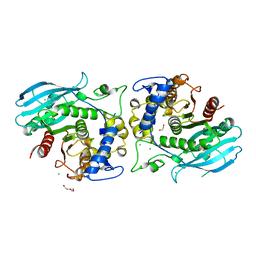 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
5H40
 
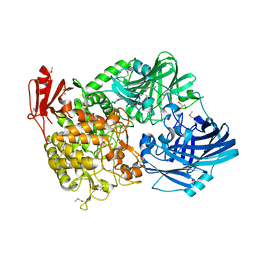 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
8J9C
 
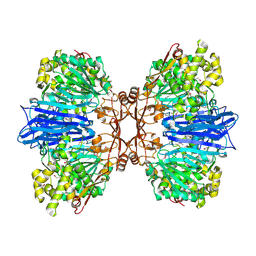 | | Crystal structure of M61 peptidase (apo-form) from Xanthomonas campestris | | Descriptor: | GLYCEROL, Putative glycyl aminopeptidase, SODIUM ION, ... | | Authors: | Yadav, P, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
6P29
 
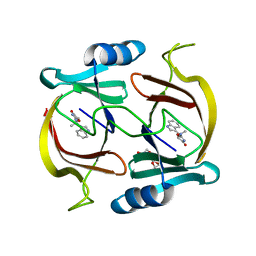 | | N-demethylindolmycin synthase (PluN2) in complex with N-demethylindolmycin | | Descriptor: | (5S)-2-amino-5-[(1R)-1-(1H-indol-3-yl)ethyl]-1,3-oxazol-4(5H)-one, N-demethylindolmycin synthase (PluN2), TRIETHYLENE GLYCOL | | Authors: | Du, Y.L, Higgins, M.A, Zhao, G, Ryan, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Convergent biosynthetic transformations to a bacterial specialized metabolite.
Nat.Chem.Biol., 15, 2019
|
|
8THE
 
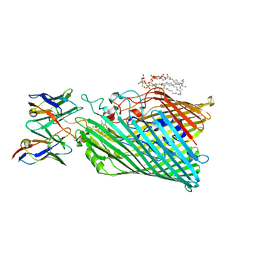 | | Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme | | Descriptor: | (2~{R},4~{R},5~{S},6~{S})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{S})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Heme/hemoglobin uptake outer membrane receptor PhuR, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Knejski, P, Erramilli, S.K, Kossiakoff, A.A. | | Deposit date: | 2023-07-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Chaperone-assisted cryo-EM structure of P. aeruginosa PhuR reveals molecular basis for heme binding.
Structure, 32, 2024
|
|
5CAA
 
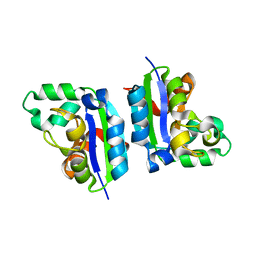 | |
8ATL
 
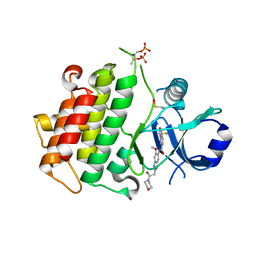 | | Discovery of IRAK4 Inhibitor 23 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[6-methoxy-2-(2-morpholin-4-yl-2-oxidanylidene-ethyl)indazol-5-yl]-6-[(1~{R})-2,2,2-tris(fluoranyl)-1-oxidanyl-ethyl]pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
7QAR
 
 | | Serial crystallography structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-methyl uric acid at room temperature | | Descriptor: | (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, Uricase | | Authors: | Bui, S, Catapano, L, Zielinski, K, Yefanov, O, Murshudov, G.N, Oberthuer, D, Steiner, R.A. | | Deposit date: | 2021-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
7QBJ
 
 | | bacterial IMPDH chimera | | Descriptor: | Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
8EAS
 
 | | Yeast VO in complex with Vma12-22p | | Descriptor: | V-type proton ATPase assembly factor Vma12p, V-type proton ATPase subunit F, V-type proton ATPase subunit a, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3AZB
 
 | | Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum in complex with NAS91-11 | | Descriptor: | 5-chloro-8-[(3-chlorobenzyl)oxy]quinoline, Beta-hydroxyacyl-ACP dehydratase, GLYCEROL | | Authors: | Maity, K, Venkata, B.S, Kapoor, N, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2011-05-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the functional and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum
J.Struct.Biol., 176, 2011
|
|
7QAB
 
 | |
6XZ3
 
 | | Crystal structure of TLNRD1 4-helix bundle | | Descriptor: | 1,2-ETHANEDIOL, Talin rod domain-containing protein 1 | | Authors: | Cowell, A, Singh, A.K, Brown, D.G, Goult, B.T. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Talin rod domain-containing protein 1 (TLNRD1) is a novel actin-bundling protein which promotes filopodia formation.
J.Cell Biol., 220, 2021
|
|
8AW1
 
 | | c-MET Y1235D mutant in complex with Tepotinib | | Descriptor: | 3-[1-(3-{5-[(1-methylpiperidin-4-yl)methoxy]pyrimidin-2-yl}benzyl)-6-oxo-1,6-dihydropyridazin-3-yl]benzonitrile, Hepatocyte growth factor receptor | | Authors: | Graedler, U, Lammens, A. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biophysical and structural characterization of the impacts of MET phosphorylation on tepotinib binding.
J.Biol.Chem., 299, 2023
|
|
5CG5
 
 | | Neutron crystal structure of human farnesyl pyrophosphate synthase in complex with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Yokoyama, T, Mizuguchi, M, Ostermann, A, Kusaka, K, Niimura, N, Schrader, T.E, Tanaka, I. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.402 Å), X-RAY DIFFRACTION | | Cite: | Protonation State and Hydration of Bisphosphonate Bound to Farnesyl Pyrophosphate Synthase
J.Med.Chem., 58, 2015
|
|
6S6Q
 
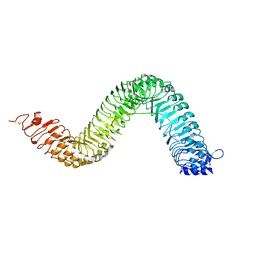 | | Crystal structure of the LRR ectodomain of the plant membrane receptor kinase GASSHO1/SCHENGEN3 from Arabidopsis thaliana in complex with CASPARIAN STRIP INTEGRITY FACTOR 2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LRR receptor-like serine/threonine-protein kinase GSO1, ... | | Authors: | Okuda, S, Moretti, A, Hothorn, M. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6S11
 
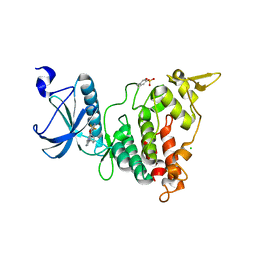 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 6-pyridin-4-yl-3-[3-(trifluoromethyloxy)phenyl]imidazo[1,2-b]pyridazine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6P62
 
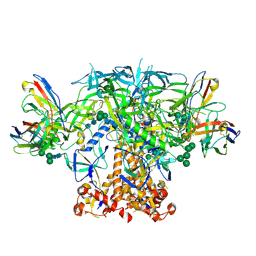 | | HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env BG505 NFL TD+, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
6S14
 
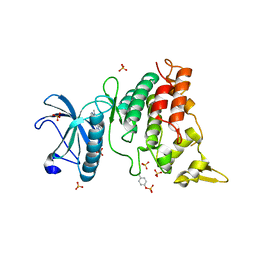 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ~{N}-cyclopropyl-~{N}-methyl-4-pyrazolo[1,5-b]pyridazin-3-yl-pyrimidin-2-amine | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6XCE
 
 | |
8FMU
 
 | | Crystal structure of human Brachyury G177D variant in complex with SJF-4601 | | Descriptor: | N-(3-chloro-4-fluorophenyl)-3-[4-(dimethylamino)butanamido]-4-methoxybenzamide, T-box transcription factor T | | Authors: | Bebenek, A, Linhares, B, Jaime-Figueroa, S, Butrin, A, Crews, C. | | Deposit date: | 2022-12-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of a Small Molecule Downmodulator for the Transcription Factor Brachyury.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3R9U
 
 | | Thioredoxin-disulfide reductase from Campylobacter jejuni. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, K.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Thioredoxin-disulfide reductase from Campylobacter jejuni.
To be Published
|
|
