6JMI
 
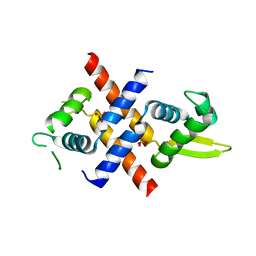 | | Crystal structure of M.tuberculosis Rv0081 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv0081 | | Authors: | Kumar, A, Phulera, S, Mande, C.S. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of hypoxic gene regulation by the Rv0081 transcription factor of Mycobacterium tuberculosis.
Febs Lett., 593, 2019
|
|
1ANX
 
 | |
7RT3
 
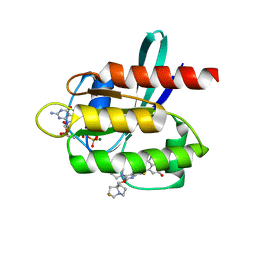 | | Crystal Structure of KRAS G12D with compound 24 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
8BN2
 
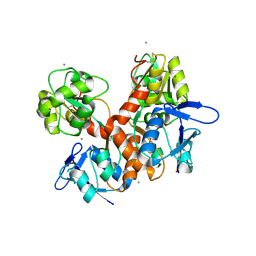 | | Crystal structure of the ligand-binding domain (LBD) of human iGluR Delta-1 (GluD1) in complex with D-Serine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Heroven, C, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | GluD1 binds GABA and controls inhibitory plasticity.
Science, 382, 2023
|
|
4ZFP
 
 | |
7RT1
 
 | | Crystal Structure of KRAS G12D with compound 15 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | Descriptor: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | Authors: | Zhou, A, Wei, Z, Bai, J, Wang, H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
7O88
 
 | |
4ZG1
 
 | |
4U2C
 
 | | Crystal structure of dienelactone hydrolase A-6 variant (S7T, A24V, Q35H, F38L, Q110L, C123S, Y145C, E199G and S208G) at 1.95 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
6FMN
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with 6-amino-2-((((2R,3S,4R,5R)-3,4-dihydroxy-5-methoxytetrahydrofuran-2-yl)methyl)amino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-azanyl-2-[[(2~{R},3~{S},4~{R},5~{R})-5-methoxy-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-3,7-dihydroimidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2018-02-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Sugar Acetonides are a Superior Motif for Addressing the Large, Solvent-Exposed Ribose-33 Pocket of tRNA-Guanine Transglycosylase.
Chemistry, 24, 2018
|
|
4U2D
 
 | | Crystal structure of dienelactone hydrolase S-2 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.67 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
7RT2
 
 | | Crystal Structure of KRAS G12D with compound 25 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GLYCEROL, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
5JI2
 
 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
6IY9
 
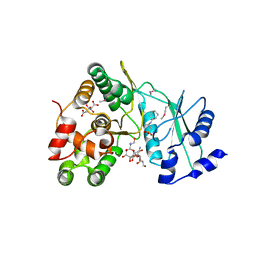 | | Crystal structure of aminoglycoside 7"-phoshotransferase-Ia (APH(7")-Ia/HYG) from Streptomyces hygroscopicus complexed with hygromycin B | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, HYGROMYCIN B VARIANT, ... | | Authors: | Takenoya, M, Shimamura, T, Yamanaka, R, Adachi, Y, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2018-12-14 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the substrate recognition of aminoglycoside 7''-phosphotransferase-Ia from Streptomyces hygroscopicus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4U2G
 
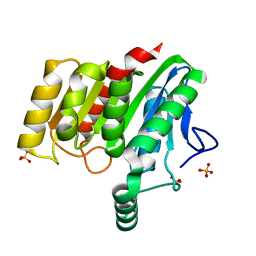 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
6IYQ
 
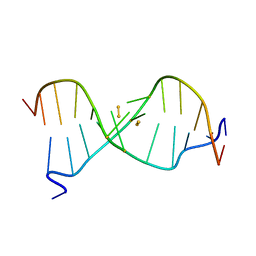 | |
7RT4
 
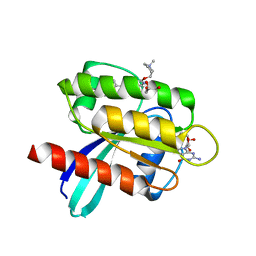 | | KRAS G12D in complex with Compound 5B (7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine) | | Descriptor: | 7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
5FAV
 
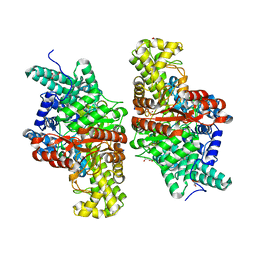 | | E491Q mutant of choline TMA-lyase | | Descriptor: | Choline trimethylamine-lyase, MALONATE ION, SODIUM ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
8BN5
 
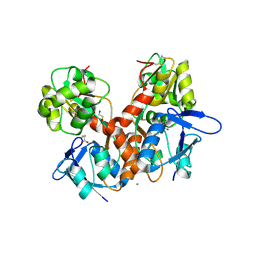 | | Crystal structure of the ligand-binding domain (LBD) of human iGluR Delta-1 (GluD1) in complex with GABA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Heroven, C, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2022-11-12 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GluD1 binds GABA and controls inhibitory plasticity.
Science, 382, 2023
|
|
1A0L
 
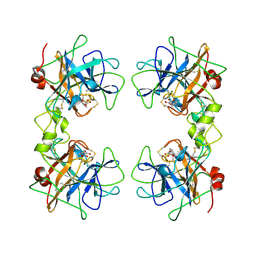 | | HUMAN BETA-TRYPTASE: A RING-LIKE TETRAMER WITH ACTIVE SITES FACING A CENTRAL PORE | | Descriptor: | (2S)-3-(4-carbamimidoylphenyl)-2-hydroxypropanoic acid, BETA-TRYPTASE | | Authors: | Pereira, P.J.B, Bergner, A, Macedo-Ribeiro, S, Huber, R, Matschiner, G, Fritz, H, Sommerhoff, C.P, Bode, W. | | Deposit date: | 1997-12-03 | | Release date: | 1999-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human beta-tryptase is a ring-like tetramer with active sites facing a central pore.
Nature, 392, 1998
|
|
5FCA
 
 | | Murine SMPDL3A in presence of excess zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
8BLJ
 
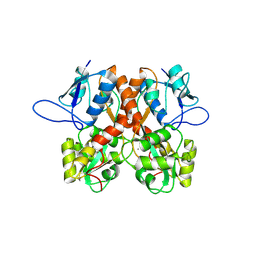 | | Crystal structure of the ligand-binding domain (LBD) of human iGluR Delta-1 (GluD1), apo state | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Heroven, C, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | GluD1 binds GABA and controls inhibitory plasticity.
Science, 382, 2023
|
|
4TWA
 
 | |
5JJN
 
 | | Structure of the SRII/HtrII(G83F) Complex in P212121 space group ("V" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
