3NR0
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R6 6/10A | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
5C4Y
 
 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
6ERQ
 
 | | Structure of the human mitochondrial transcription initiation complex at the HSP promoter | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
7OF0
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
1K3E
 
 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
3A8O
 
 | | Crystal structure of Nitrile Hydratase complexed with Trimethylacetamide | | Descriptor: | 2,2-dimethylpropanamide, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
6E5V
 
 | | human mGlu8 receptor amino terminal domain in complex with (S)-3,4-Dicarboxyphenylglycine (DCPG) | | Descriptor: | 4-[(S)-amino(carboxy)methyl]benzene-1,2-dicarboxylic acid, CHLORIDE ION, Metabotropic glutamate receptor 8 | | Authors: | Chen, Q, Ho, J.D, Ashok, S, Vargas, M.C, Wang, J, Atwell, S, Bures, M, Schkeryantz, J.M, Monn, J.A, Hao, J. | | Deposit date: | 2018-07-23 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for ( S)-3,4-Dicarboxyphenylglycine (DCPG) As a Potent and Subtype Selective Agonist of the mGlu8Receptor.
J. Med. Chem., 61, 2018
|
|
1TQJ
 
 | | Crystal structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 angstrom resolution | | Descriptor: | Ribulose-phosphate 3-epimerase | | Authors: | Wise, E.L, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-17 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6YTY
 
 | | CLK3 A319V mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
4V8Z
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
1K4Q
 
 | | Human Glutathione Reductase Inactivated by Peroxynitrite | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione Reductase | | Authors: | Savvides, S.N, Scheiwein, M, Boehme, C.C, Arteel, G.E, Karplus, P.A, Becker, K, Schirmer, R.H. | | Deposit date: | 2001-10-08 | | Release date: | 2002-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the antioxidant enzyme glutathione reductase inactivated by peroxynitrite.
J.Biol.Chem., 277, 2002
|
|
4D6Y
 
 | | Crystal structure of the receiver domain of NtrX from Brucella abortus in complex with beryllofluoride and magnesium | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Otero, L.H, Fernandez, I, Carrica, M.C, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
4CH2
 
 | | Low-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
2ONC
 
 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
4CJM
 
 | | Crystal structure of human FGF18 | | Descriptor: | FIBROBLAST GROWTH FACTOR 18, SULFATE ION | | Authors: | Brown, A, Adam, L.E, Blundell, T.L. | | Deposit date: | 2013-12-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Fibroblast Growth Factor 18 (Fgf18)
Protein Cell, 5, 2014
|
|
4M3F
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | HTH-type transcriptional regulator EthR, N-(3-methylbutyl)-4-(2-methyl-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
7RWU
 
 | |
7O5H
 
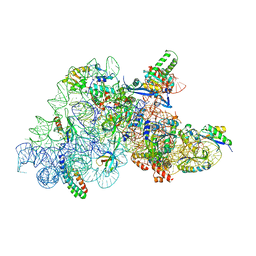 | | Ribosomal methyltransferase KsgA bound to small ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Stephan, N.C, Ries, A.B, Boehringer, D, Ban, N. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA.
Nucleic Acids Res., 49, 2021
|
|
4CN7
 
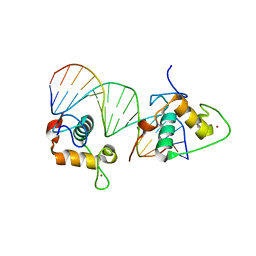 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to an idealized DR1 Response Element | | Descriptor: | 5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP *AP*GP)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP *AP*GP)-3', CHLORIDE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
1U58
 
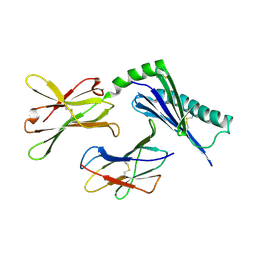 | | Crystal structure of the murine cytomegalovirus MHC-I homolog m144 | | Descriptor: | MHC-I homolog m144, beta-2-microglobulin | | Authors: | Natarajan, K, Hicks, A, Robinson, H, Guan, R, Margulies, D.H. | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the murine cytomegalovirus MHC-I homolog m144.
J.Mol.Biol., 358, 2006
|
|
2D1U
 
 | |
6QJF
 
 | |
5OWE
 
 | |
5XA0
 
 | | Crystal structure of inositol 1,4,5-trisphosphate receptor cytosolic domain | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Hamada, K, Miyatake, H, Terauchi, A, Mikoshiba, K. | | Deposit date: | 2017-03-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (5.812 Å) | | Cite: | IP3-mediated gating mechanism of the IP3 receptor revealed by mutagenesis and X-ray crystallography
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
