4R6E
 
 | | Human artd1 (parp1) - catalytic domain in complex with inhibitor niraparib | | Descriptor: | 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Karlberg, T, Thorsell, A.G, Brock, J, Schuler, H. | | Deposit date: | 2014-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J.Med.Chem., 60, 2017
|
|
3LYA
 
 | |
8GBL
 
 | | Structure of Apo Human SIRT5 | | Descriptor: | NAD-dependent protein deacylase sirtuin-5, mitochondrial, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
6PAJ
 
 | |
6SAY
 
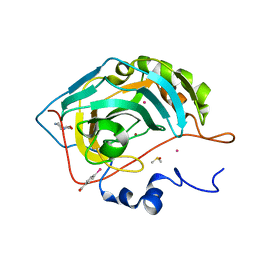 | | Human Carbonic Anhydrase II in complex with fragment. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, (8~{a}~{S})-1,5,6,7,8,8~{a}-hexahydro-[1,3]oxazolo[3,4-a]pyrazin-3-one, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-07-18 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
8BOY
 
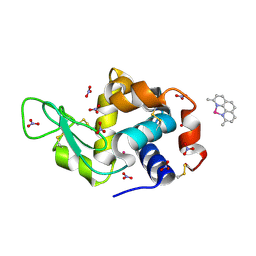 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 4.0 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], Lysozyme C, NITRATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
6YNN
 
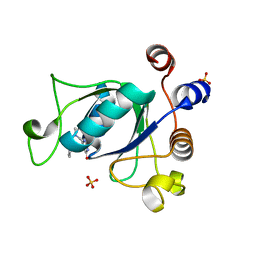 | | Crystal structure of YTHDC1 with compound DHU_DC1_135 | | Descriptor: | 6-[[(2-chloranyl-6-fluoranyl-phenyl)methyl-methyl-amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
4R9L
 
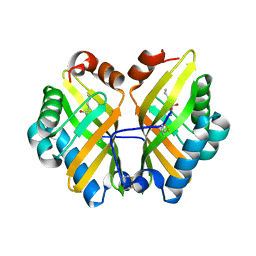 | | Structure of a thermostable elevenfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis, containing two stabilizing disulfide bonds | | Descriptor: | (2R)-2-hydroxyhexanamide, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
7RTD
 
 | |
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | Authors: | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-08 | | Release date: | 2014-09-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8515 Å) | | Cite: | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
5MGG
 
 | |
7S7V
 
 | | Crystal structure of iNicSnFR3a Fluorescent Nicotine Sensor | | Descriptor: | iNicSnFR 3.0 Fluorescent Nicotine Sensor | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Correction: Fluorescence activation mechanism and imaging of drug permeation with new sensors for smoking-cessation ligands.
Elife, 11, 2022
|
|
6YOQ
 
 | |
5MGL
 
 | |
4QQE
 
 | | Crystal structure of WDR5, WD repeat domain 5 in complex with compound SGC-DS-MT-0345 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[2-(4-methylpiperazin-1-yl)-5-(quinolin-3-yl)phenyl]-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Wernimont, A, Smil, D, Getlik, M, Senisterra, G, Poda, G, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of WDR5, WD repeat domain 5 in complex with compound SGC-DS-MT-0345
To be Published
|
|
6PDP
 
 | | Human PIM1 bound to benzothiophene inhibitor 379 | | Descriptor: | 5-[2-(acetylamino)-1-benzothiophen-4-yl]-N-cyclopropylthiophene-2-carboxamide, Peptide, SULFATE ION, ... | | Authors: | Godoi, P.H.C, Sriranganadane, D, Santiago, A.S, Fala, A.M, Ramos, P.Z, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human PIM1
To Be Published
|
|
8BDL
 
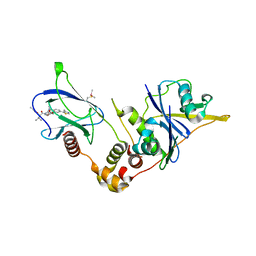 | | VCB in complex with compound 27 | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-1-[2-(2-methoxyethoxy)-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8BDM
 
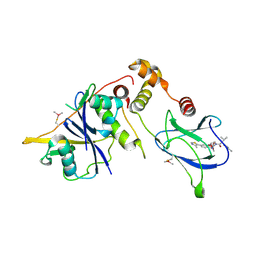 | | VCB in complex with compound 26 | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-(2-methoxyethoxy)-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
7MVX
 
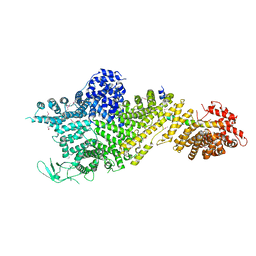 | | Crystal structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5MFB
 
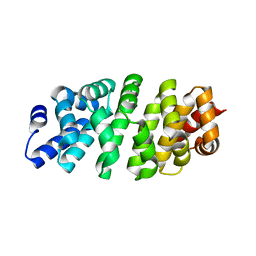 | | Designed armadillo repeat protein YIII(Dq)4CqI | | Descriptor: | YIII(Dq)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
3LTN
 
 | | Inhibitor-stabilized topoisomerase IV-DNA cleavage complex (S. pneumoniae) | | Descriptor: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2010-02-16 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
8G4K
 
 | |
5JS9
 
 | | Uncleaved prefusion optimized gp140 trimer with an engineered 8-residue HR1 turn bound to broadly neutralizing antibodies 8ANC195 and PGT128 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (6.918 Å) | | Cite: | Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability.
Nat Commun, 7, 2016
|
|
6YPZ
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
