4UNQ
 
 | | Mtb TMK in complex with compound 36 | | Descriptor: | 4-[(R)-methylsulfinyl]-2-oxo-6-[3-(trifluoromethoxy)phenyl]-1,2-dihydropyridine-3-carbonitrile, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
7Q0Y
 
 | | Crystal structure of CTX-M-14 in complex with Bortezomib | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
4UNS
 
 | | Mtb TMK in complex with compound 40 | | Descriptor: | N-[4-(3-CYANO-7-ETHYL-5-METHYL-2-OXO-1H-1,6-NAPHTHYRIDIN-4-YL)PHENYL]METHANESULFONAMIDE, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
7Q4I
 
 | | Crystal structure of DmC1GalT1 in complex with UDP-Mn2+ and the APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1, ... | | Authors: | Gonzalez-Ramirez, A.M, Coelho, H, Companon, I, Grosso, A.S, Yang, Z, Narimatsu, Y, Clausen, H, Marcelo, F, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2021-10-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the synthesis of the core 1 structure by C1GalT1.
Nat Commun, 13, 2022
|
|
7PWY
 
 | | Structure of human dimeric ACMSD in complex with the inhibitor TES-1025 | | Descriptor: | 2-[3-[(5-cyano-6-oxidanylidene-4-thiophen-2-yl-1H-pyrimidin-2-yl)sulfanylmethyl]phenyl]ethanoic acid, 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, POTASSIUM ION, ... | | Authors: | Cianci, M, Giacche, N, Carotti, A, Liscio, P, Amici, A, Cialabrini, L, De Franco, F, Pellicciari, R, Raffaelli, N. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Human Dimeric alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Inhibition With TES-1025.
Front Mol Biosci, 9, 2022
|
|
4UMI
 
 | |
4UMP
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-(isoquinolin-7-yl)prop-2-yn-1-ol, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
7PRR
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
4UW8
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber with its intra-molecular chaperone domain | | Descriptor: | CITRATE ANION, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4US3
 
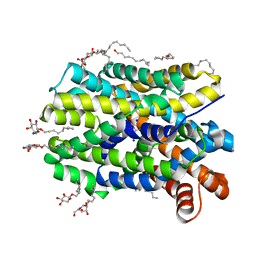 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
4V73
 
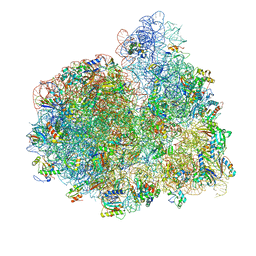 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre5a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7PX8
 
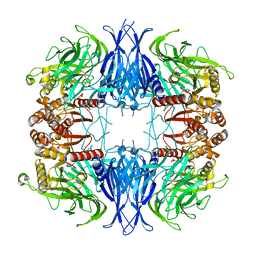 | | CryoEM structure of mammalian acylaminoacyl-peptidase | | Descriptor: | Acylamino-acid-releasing enzyme | | Authors: | Kiss-Szeman, A.J, Harmat, V, Menyhard, D.K, Straner, P, Jakli, I, Hosogi, N, Perczel, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structure of acylpeptide hydrolase reveals substrate selection by multimerization and a multi-state serine-protease triad.
Chem Sci, 13, 2022
|
|
4V8S
 
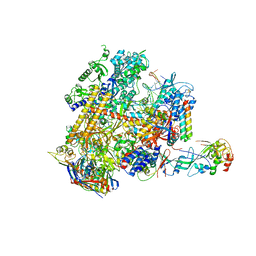 | | Archaeal RNAP-DNA binary complex at 4.32Ang | | Descriptor: | 5'-D(*AP*TP*AP*GP*AP*GP*TP*AP*TP*AP*AP*GP*AP*TP *AP*G)-3', 5'-D(*TP*CP*TP*TP*AP*TP*AP*CP*TP*CP*TP*AP*TP*CP)-3', DNA-DIRECTED RNA POLYMERASE, ... | | Authors: | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2012-07-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.323 Å) | | Cite: | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
4USP
 
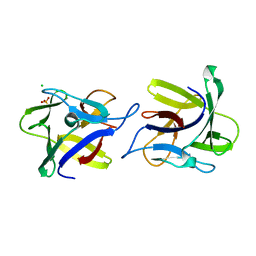 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
4V71
 
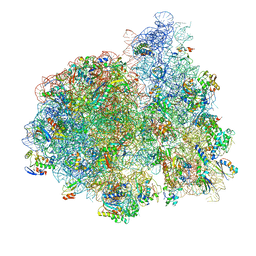 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate pre-translocation state (pre2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6WQA
 
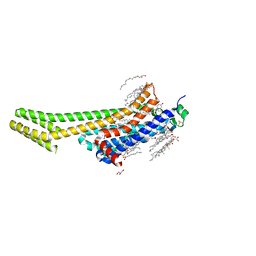 | | 2.0A angstrom A2a adenosine receptor structure using XFEL data collected in helium atmosphere. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Lee, M.-Y, Geiger, J, Ishchenko, A, Han, G.W, Barty, A, White, T.A, Gati, C, Batyuk, A, Hunter, M.S, Aquila, A, Boutet, S, Weierstall, U, Cherezov, V, Liu, W. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Harnessing the power of an X-ray laser for serial crystallography of membrane proteins crystallized in lipidic cubic phase
Iucrj, 7, 2020
|
|
7Q1V
 
 | |
6YRD
 
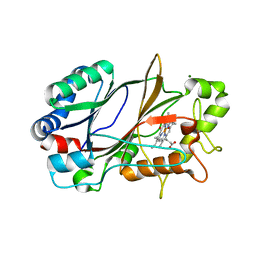 | | SFX structure of dye-type peroxidase DtpB in the ferryl state | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Axford, D.A, Owen, R.L, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7PH7
 
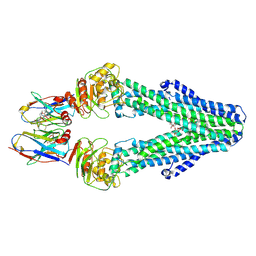 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position T68C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-binding transport protein multicopy suppressor of htrB, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PPM
 
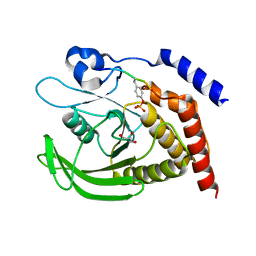 | | SHP2 catalytic domain in complex with IRS1 (889-901) phosphopeptide (pSer-892, pTyr-896) | | Descriptor: | GLYCEROL, Insulin receptor substrate 1, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
6Y1K
 
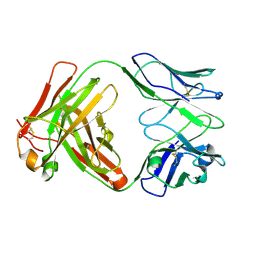 | | Crystal structure of the unmodified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | FAB A.17 L47R mutant Heavy Chain, FAB A.17 L47R mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YBL
 
 | | Structure of MBP-Mcl-1 in complex with compound 9m | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6YRC
 
 | | Spectroscopically-validated structure of DtpB from Streptomyces lividans in the ferric state | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lucic, M, Dworkowski, F.S.N, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YBJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 3e | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[(2-methylpyrazol-3-yl)methoxy]phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6Y1M
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47K mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47K mutant HEAVY CHAIN, FAB A.17 L47K mutant Light CHAIN | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
