5PB8
 
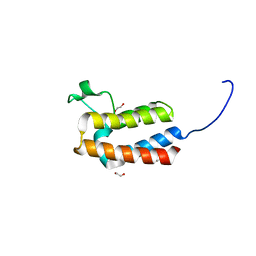 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B in complex with N09522a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, P-HYDROXYACETOPHENONE | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5ARG
 
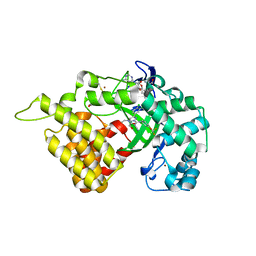 | | SMYD2 in complex with SGC probe BAY-598 | | Descriptor: | GLYCEROL, N-LYSINE METHYLTRANSFERASE SMYD2, N-[1-(N'-CYANO-N-[3-(DIFLUOROMETHOXY)PHENYL]CARBAMIMIDOYL)-3-(3,4-DICHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-4-YL]-N-ETHYL-2-HYDROXYACETAMIDE, ... | | Authors: | Hillig, R.C, Badock, V, Barak, N, Stellfeld, T, Eggert, E, ter Laak, A, Weiske, J, Christ, C.D, Koehr, S, Stoeckigt, D, Mowat, J, Mueller, T, Fernandez-Montalvan, A.E, Hartung, I.V, Stresemann, C, Brumby, T, Weinmann, H. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (Bay-598) for the Protein Lysine Methyltransferase Smyd2.
J.Med.Chem., 59, 2016
|
|
5AP3
 
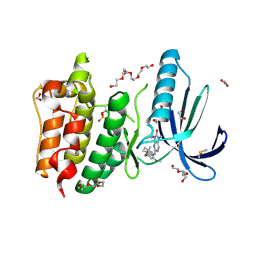 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 9-CYCLOPENTYL-2-[[2-METHOXY-4-[(1-METHYLPIPERIDIN-4-YL)OXY]-PHENYL]AMINO]-7-METHYL-7,9-DIHYDRO-8H-PURIN-8-ONE, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
5AR7
 
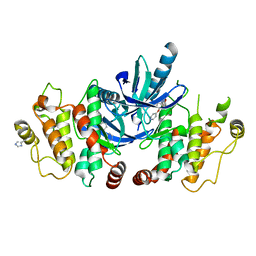 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biaryl Urea | | Descriptor: | 1-(5-TERT-BUTYL-1,2-OXAZOL-3-YL)-3-(4-PYRIDIN-4-YLOXYPHENYL)UREA, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
3NJ6
 
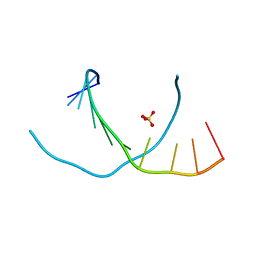 | | 0.95 A resolution X-ray structure of (GGCAGCAGCC)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*CP*AP*GP*CP*C)-3', SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases.
Nucleic Acids Res., 38, 2010
|
|
8QSO
 
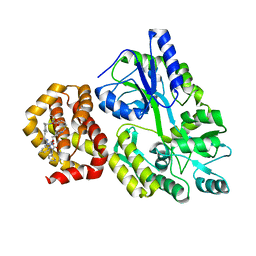 | | Crystal structure of human Mcl-1 in complex with compound 1 | | Descriptor: | (13S,16R,19S)-16-benzyl-43-ethoxy-N-methyl-7,11,14,17-tetraoxo-13-phenyl-5-oxa-2,8,12,15,18-pentaaza-1(1,4),4(1,2)-dibenzena-9(1,4)-cyclohexanacycloicosaphane-19-carboxamide, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hekking, K.F.W, Gremmen, S, Maroto, S, Keefe, A.D, Zhang, Y. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Development of Potent Mcl-1 Inhibitors: Structural Investigations on Macrocycles Originating from a DNA-Encoded Chemical Library Screen.
J.Med.Chem., 67, 2024
|
|
5AQO
 
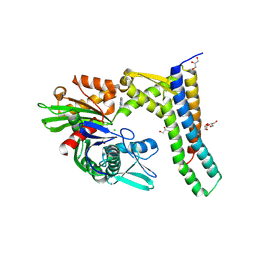 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-METHYLQUINAZOLIN-4-AMINE, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AMH
 
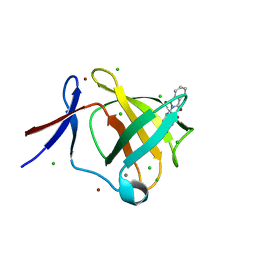 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, trigonal crystal form | | Descriptor: | CALCIUM ION, CEREBLON ISOFORM 4, CHLORIDE ION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5PBA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B in complex with N09460a | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-hydroxypyridine-2-carboxylic acid, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5AQG
 
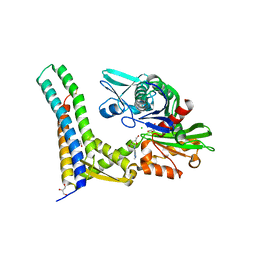 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (2R,3R,4S,5R)-2-(3-AMINO-5-METHYL-1,4,5,6,8-PENTAAZAACENAPHTHYLEN-1(5H)-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
6WLA
 
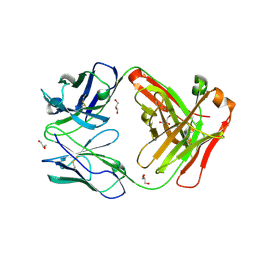 | | Antigen binding fragment of ch128.1 | | Descriptor: | Fab ch128.1 heavy chain, Fab ch128.1 light chain, GLYCEROL | | Authors: | Helguera, G, Rodriguez, J.A, Sawaya, M, Cascio, D, Zink, S, Ziegenbein, J, Short, C. | | Deposit date: | 2020-04-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Host receptor-targeted therapeutic approach to counter pathogenic New World mammarenavirus infections.
Nat Commun, 13, 2022
|
|
3NF0
 
 | | mPlum-TTN | | Descriptor: | D-MALATE, Fluorescent protein plum | | Authors: | Mayo, S.L, Chica, R.A, Moore, M.M. | | Deposit date: | 2010-06-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Changes in the Fluorescent Properties of Computationally Designed Red Fluorescent Proteins
To be Published
|
|
6KPC
 
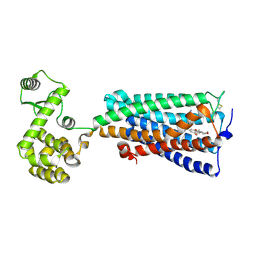 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
5BQI
 
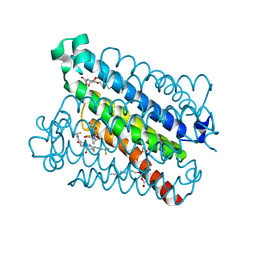 | | Discovery of a Potent and Selective mPGES-1 Inhibitor for the Treatment of Pain | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-(difluoromethyl)-5-{[(2-methylpropanoyl)amino]methyl}-N-{5-methyl-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-2-yl}pyridine-3-carboxamide, GLUTATHIONE, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
3NJ7
 
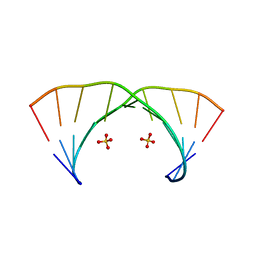 | | 1.9 A resolution X-ray structure of (GGCAGCAGCC)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*CP*AP*GP*CP*C)-3', SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases.
Nucleic Acids Res., 38, 2010
|
|
2DW6
 
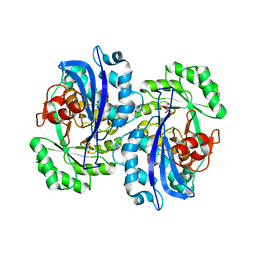 | | Crystal structure of the mutant K184A of D-Tartrate Dehydratase from Bradyrhizobium japonicum complexed with Mg++ and D-tartrate | | Descriptor: | Bll6730 protein, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
5B5H
 
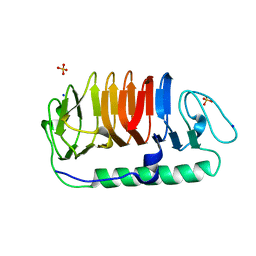 | | Hydrophobic ice-binding site confer hyperactivity on antifreeze protein from a snow mold fungus | | Descriptor: | Antifreeze protein, SODIUM ION, SULFATE ION | | Authors: | Cheng, J, Hanada, Y, Miura, A, Tsuda, S, Kondo, H. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Hydrophobic ice-binding sites confer hyperactivity of an antifreeze protein from a snow mold fungus.
Biochem.J., 473, 2016
|
|
6KXE
 
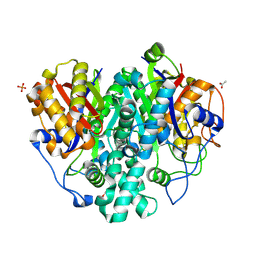 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
5B5Q
 
 | |
2DSU
 
 | | Binding of chitin-like polysaccharide to protective signalling factor: Crystal structure of the complex formed between signalling protein from sheep (SPS-40) with a tetrasaccharide at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
5BK0
 
 | | Crystal structure of 663 Fab bound to circumsporozoite protein NANP 5-mer | | Descriptor: | 663 Antibody, heavy chain, light chain, ... | | Authors: | Scally, S.W, Bosch, A, Triller, G, Wardemann, H, Julien, J.P. | | Deposit date: | 2017-09-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies.
Immunity, 47, 2017
|
|
7SPM
 
 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (30 mg/mL EDC, 12 hours, 2 doses). | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
5PBC
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B in complex with N09724a | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1H-imidazole, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3NQ1
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium in complex with inhibitor kojic acid | | Descriptor: | 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
3NQ5
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium R209H mutant | | Descriptor: | COPPER (II) ION, Tyrosinase, ZINC ION | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
