6NMT
 
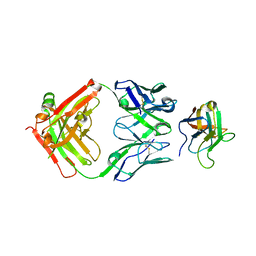 | | Non-Blocking Fab 3 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 3 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 3 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NN4
 
 | | The structure of human liver pyruvate kinase, hLPYK-D499N, in complex with Fru-1,6-BP | | 分子名称: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, PHOSPHOENOLPYRUVATE, ... | | 著者 | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | 登録日 | 2019-01-14 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NDP
 
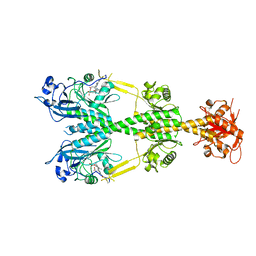 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant L193Q from Xanthomonas campestris | | 分子名称: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | 著者 | Otero, L.H, Sirigu, S, Klinke, S, Rinaldi, J, Conforte, V, Malamud, F, Goldbaum, F.A, Chavas, L, Bonomi, H.R. | | 登録日 | 2018-12-14 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.89 Å) | | 主引用文献 | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
6XHX
 
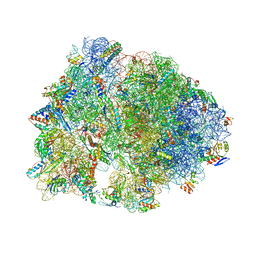 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with erythromycin and protein Y (YfiA) at 2.55A resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | 登録日 | 2020-06-19 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
8G1U
 
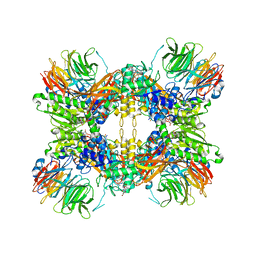 | | Structure of the methylosome-Lsm10/11 complex | | 分子名称: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | 著者 | Lin, M, Paige, A, Tong, L. | | 登録日 | 2023-02-03 | | 公開日 | 2023-08-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
8DBX
 
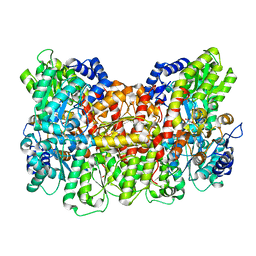 | |
6XCF
 
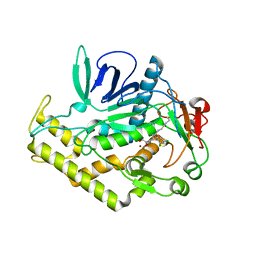 | |
6NJ2
 
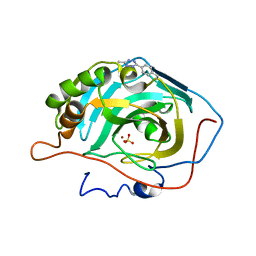 | |
6X9C
 
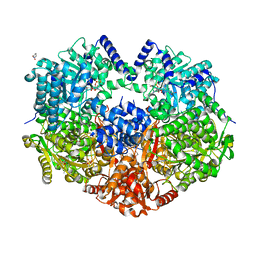 | |
6QAZ
 
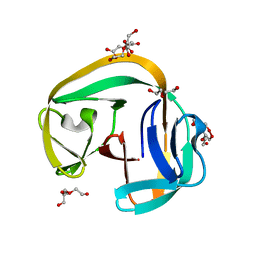 | | Crystal structure of gp41-1 intein | | 分子名称: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | 著者 | Mikula, K.M, Beyer, H.M, Li, M, Wlodawer, A, Iwai, H. | | 登録日 | 2018-12-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | The crystal structure of the naturally split gp41-1 intein guides the engineering of orthogonal split inteins from cis-splicing inteins.
Febs J., 287, 2020
|
|
6NKL
 
 | | 2.2 A resolution structure of VapBC-1 from nontypeable Haemophilus influenzae | | 分子名称: | Antitoxin VapB1, Ribonuclease VapC | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Molinaro, A.L, Daines, D.A. | | 登録日 | 2019-01-07 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of VapBC-1 from Nontypeable Haemophilus influenzae and the Effect of PIN Domain Mutations on Survival during Infection.
J.Bacteriol., 201, 2019
|
|
8D0Z
 
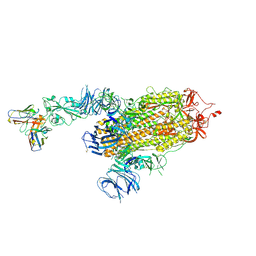 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | 著者 | Ozorowski, G, Torres, J.L, Ward, A.B. | | 登録日 | 2022-05-26 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
6XCL
 
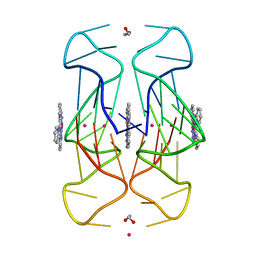 | | Crystal Structure of human telomeric DNA G-quadruplex in complex with a novel platinum(II) complex. | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), POTASSIUM ION, ... | | 著者 | Miron, C.E, van Staalduinen, L.M, Jia, Z, Petitjean, A. | | 登録日 | 2020-06-08 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Going Platinum to the Tune of a Remarkable Guanine Quadruplex Binder: Solution- and Solid-State Investigations.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6PT8
 
 | | Crystal Structure of CobT from Methanocaldococcus jannaschii in complex with Adenine Alpha-Ribotide and Nicotinic Acid | | 分子名称: | ALPHA-ADENOSINE MONOPHOSPHATE, NICOTINIC ACID, PHOSPHATE ION, ... | | 著者 | Schwarzwalder, A.H, Jeter, V.L, Vecellio, A.A, Erpenbach, E, Escalante, J.C, Rayment, I. | | 登録日 | 2019-07-15 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural studies of the phosphoribosyltransferase involved in cobamide biosynthesis in methanogenic archaea and cyanobacteria.
Sci Rep, 12, 2022
|
|
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6Q94
 
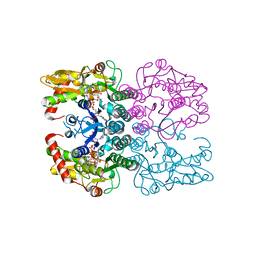 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | 分子名称: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | 著者 | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2018-12-17 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
7S8R
 
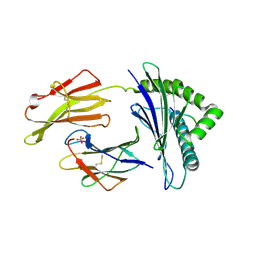 | | Crystal Structure of HLA A*1101 in complex with SALEWIKNK, an 9-mer epitope from Influenza B | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | 著者 | Nguyen, A.T, Szeto, C, Gras, S. | | 登録日 | 2021-09-19 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
6NMS
 
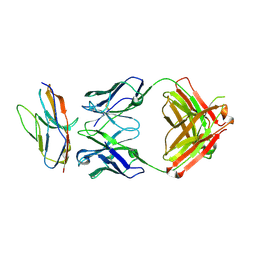 | | Blocking Fab 136 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 136 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 136 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6PUH
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-Ribityl-less | | 分子名称: | 6-methyl-5-[(1E)-3-oxobut-1-en-1-yl]pyrimidine-2,4(1H,3H)-dione, Beta-2-microglobulin, Human TCR alpha chain, ... | | 著者 | Awad, W, Keller, A.N, Rossjohn, J. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
7S8Q
 
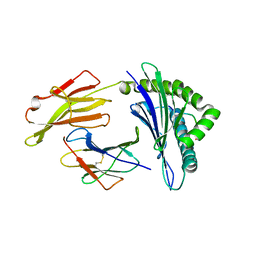 | | Crystal Structure of HLA A*1101 in complex with KTNGNAFIGK, an 10-mer epitope from Influenza B | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | 著者 | Nguyen, A.T, Szeto, C, Gras, S. | | 登録日 | 2021-09-19 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
8DK6
 
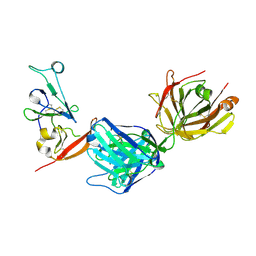 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | 著者 | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | 登録日 | 2022-07-03 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
7A78
 
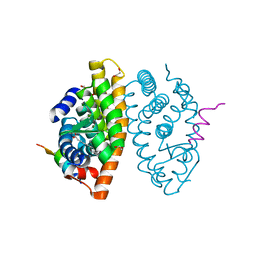 | | Crystal structure of RXR beta LBD in complexes with palmitic acid and GRIP-1 peptide | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | 著者 | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
5N1Y
 
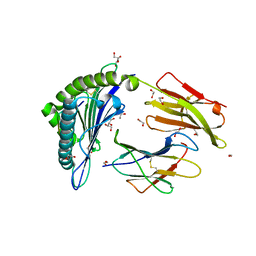 | | HLA-A02 carrying MVWGPDPLYV | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | 登録日 | 2017-02-06 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J. Clin. Invest., 126, 2016
|
|
6WAX
 
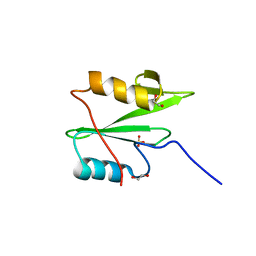 | | C-terminal SH2 domain of p120RasGAP | | 分子名称: | 1,2-ETHANEDIOL, Ras GTPase-activating protein 1, SULFATE ION | | 著者 | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
7ACD
 
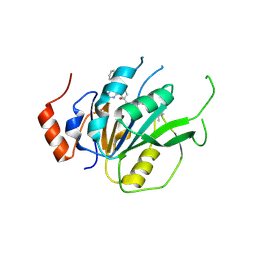 | | Crystal structure of the human METTL3-METTL14 complex with compound T30 (UZH1a) | | 分子名称: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-oxidanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Bedi, R.K, Huang, D, Caflisch, A. | | 登録日 | 2020-09-10 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes.
Chemmedchem, 16, 2021
|
|
