5KVY
 
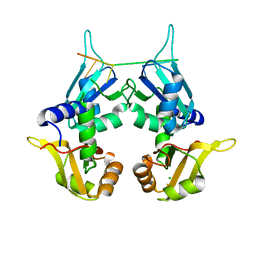 | | CRYSTAL STRUCTURE OF THE TWO TANDEM RRM DOMAINS OF PUF60 BOUND TO A PORTION OF AN ADML PRE-MRNA 3' SPLICE SITE ANALOG | | 分子名称: | CHLORIDE ION, DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | 著者 | Hsiao, H.-H, Crichlow, G.V, Albright, R.A, Murphy, J.W, Lolis, E.J, Braddock, D.T. | | 登録日 | 2016-07-15 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
6ETN
 
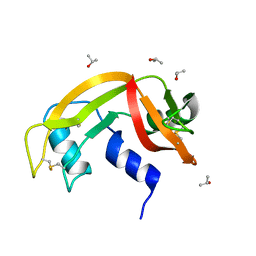 | |
8B3L
 
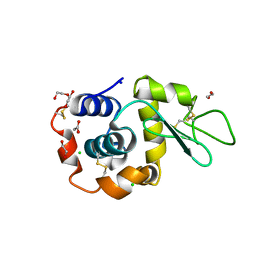 | | Hen Egg White Lysozyme 2s in situ crystallization | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | 登録日 | 2022-09-16 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
1KD1
 
 | | Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui | | 分子名称: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | 著者 | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | 登録日 | 2001-11-12 | | 公開日 | 2002-07-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
4YD4
 
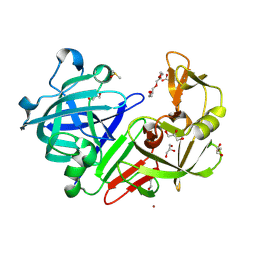 | |
5L76
 
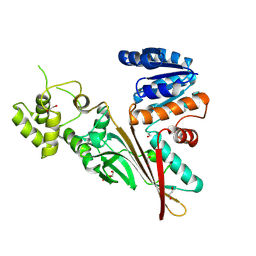 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in apo form) | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | 著者 | Kopec, J, Pena, I.A, Rembeza, E, Strain-Damerell, C, Chalk, R, Borkowska, O, Goubin, S, Velupillai, S, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Arruda, P, Yue, W.W. | | 登録日 | 2016-06-02 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in apo form)
To Be Published
|
|
8BC7
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex an aminoglutarimide degron peptide | | 分子名称: | Cereblon isoform 4, PHE-PHE-GLU-GLN-MET-GLN-QCI, S-Thalidomide, ... | | 著者 | Heim, C, Albrecht, R, Spring, A.K, Hartmann, M.D. | | 登録日 | 2022-10-15 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.719 Å) | | 主引用文献 | Identification and structural basis of C-terminal cyclic imides as natural degrons for cereblon.
Biochem.Biophys.Res.Commun., 637, 2022
|
|
6VB1
 
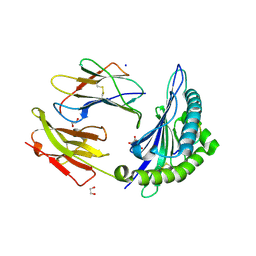 | | HLA-B*15:02 complexed with a synthetic peptide | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Schutte, R.J, Li, D, Andring, J, McKenna, R, Ostrov, D.A. | | 登録日 | 2019-12-18 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | HLA-B*15:02 complexed with a synthetic peptide
To Be Published
|
|
8F1U
 
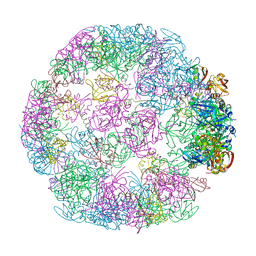 | | Structure of a 24mer DegP cage bound to the client protein hTRF1 | | 分子名称: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | 著者 | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | 登録日 | 2022-11-06 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (13.8 Å) | | 主引用文献 | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
6HK6
 
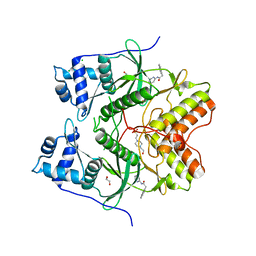 | | Human RIOK2 bound to inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-naphthalen-2-yl-~{N}-pyridin-2-yl-ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Wang, J, Krojer, T, Bountra, C, Edwards, A.M, Arrowsmith, C, Knapp, S, Elkins, J.M. | | 登録日 | 2018-09-05 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of human RIOK2 bound to a specific inhibitor.
Open Biology, 9, 2019
|
|
4TKX
 
 | | Structure of Protease | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, LEAD (II) ION, ... | | 著者 | Gorman, M.A, Parker, M.W. | | 登録日 | 2014-05-28 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the lysine specific protease Kgp from Porphyromonas gingivalis, a target for improved oral health.
Protein Sci., 24, 2015
|
|
5L7K
 
 | |
5KXT
 
 | | Hen Egg White Lysozyme at 278K, Data set 5 | | 分子名称: | Lysozyme C, SODIUM ION | | 著者 | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | 登録日 | 2016-07-20 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5ADO
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - Light chain S35R mutant | | 分子名称: | DIETHYL PHOSPHONATE, FAB A.17 | | 著者 | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | 登録日 | 2015-08-21 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
6EUR
 
 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Fe(II)/alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | 著者 | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | 登録日 | 2017-10-31 | | 公開日 | 2018-11-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
5L88
 
 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSILATED, CRYSTAL FORM I, non-parsimonious model | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Anti-afamin antibody N14, Fab fragment, ... | | 著者 | Rupp, B, Naschberger, A. | | 登録日 | 2016-06-07 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6B7Z
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | 分子名称: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2018-01-10 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5NAL
 
 | | The crystal structure of inhibitor-15 covalently bound to PDE6D | | 分子名称: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}4-[(4-chlorophenyl)methyl]-~{N}1-(cyclohexylmethyl)-~{N}4-cyclopentyl-~{N}1-[(~{Z})-4-[(~{E})-methyliminomethyl]-5-oxidanyl-hex-4-enyl]benzene-1,4-disulfonamide | | 著者 | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | 登録日 | 2017-02-28 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Covalent Protein Labeling at Glutamic Acids.
Cell Chem Biol, 24, 2017
|
|
6M96
 
 | |
8FX4
 
 | | GC-C-Hsp90-Cdc37 regulatory complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Guanylyl cyclase C, Heat shock protein HSP 90-beta, ... | | 著者 | Caveney, N.A, Garcia, K.C. | | 登録日 | 2023-01-23 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insight into guanylyl cyclase receptor hijacking of the kinase-Hsp90 regulatory mechanism.
Elife, 12, 2023
|
|
8V47
 
 | |
8V46
 
 | |
6FC4
 
 | |
6B9R
 
 | |
7JIW
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder530 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
