5MY7
 
 | | Adhesin Complex Protein from Neisseria meningitidis | | Descriptor: | Adhesin, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Derrick, J.P, Awanye, A. | | Deposit date: | 2017-01-25 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Neisseria Adhesin Complex Protein (ACP) and its role as a novel lysozyme inhibitor.
PLoS Pathog., 13, 2017
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-26 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
6P6G
 
 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7UDP
 
 | | Crystal structure of COQ8A-CA157 inhibitor complex in space group C2 | | Descriptor: | 4-[(3,4,5-trimethoxyphenyl)amino]quinoline-7-carbonitrile, Atypical kinase COQ8A, mitochondrial | | Authors: | Bingman, C.A, Murray, N, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Small-molecule inhibition of the archetypal UbiB protein COQ8.
Nat.Chem.Biol., 19, 2023
|
|
8AJ2
 
 | |
4YR7
 
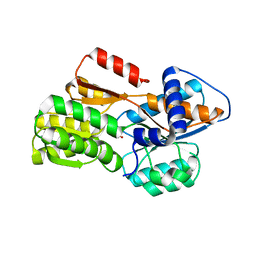 | |
8ACB
 
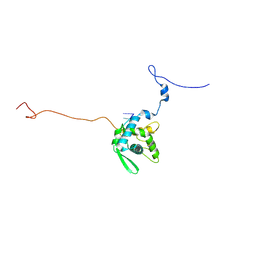 | | CryoEM structure of sweet potato feathery mottle virus VLP | | Descriptor: | Genome polyprotein, Single-stranded RNA | | Authors: | Javed, A, Byrne, J.M, Ranson, N, Lomonosoff, G. | | Deposit date: | 2022-07-05 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host.
Commun Biol, 6, 2023
|
|
7NG7
 
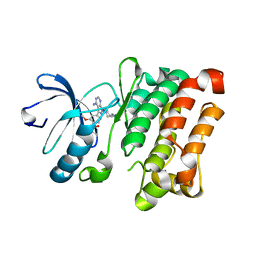 | | Src kinase bound to eCF506 trapped in inactive conformation | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Src, tert-butyl (4-(4-amino-1-(2-(4-(dimethylamino)piperidin-1-yl)ethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-2-methoxyphenyl)carbamate | | Authors: | Lietha, D, Unciti-Broceta, A. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Conformation Selective Mode of Inhibiting SRC Improves Drug Efficacy and Tolerability.
Cancer Res., 81, 2021
|
|
2UWC
 
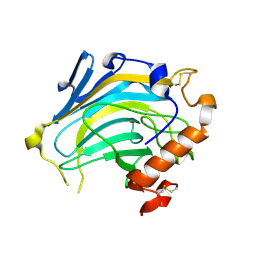 | | Crystal structure of Nasturtium xyloglucan hydrolase isoform NXG2 | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J.M, Michel, G, Kallas, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
8ACC
 
 | | CryoEM structure of sweet potato mild mottle virus VLP | | Descriptor: | Polyprotein, Single-stranded RNA | | Authors: | Javed, A, Byrne, B.M, Ranson, N, Lomonosoff, G. | | Deposit date: | 2022-07-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host.
Commun Biol, 6, 2023
|
|
4R6K
 
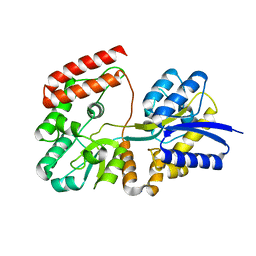 | | Crystal structure of ABC transporter substrate-binding protein YesO from Bacillus subtilis, TARGET EFI-510761, an open conformation | | Descriptor: | SODIUM ION, SOLUTE-BINDING PROTEIN | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of transporter Yeso from Bacillus subtilis, Target Efi-510761
To be Published
|
|
5MAZ
 
 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PA14 IN COMPLEX WITH 3-Thiophenesulfonamide-2,5-dimethyl-N-methyl-beta-L-fucopyranoside | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Sommer, R, Imberty, A, Titz, A, Varrot, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of a new class of C-glycoside based inhibitors of the virulence factor LecB from Pseudomonas aeruginosa
To Be Published
|
|
3OT5
 
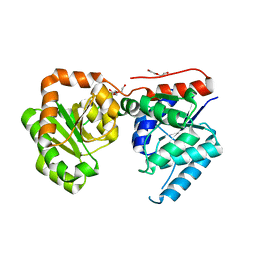 | | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes.
TO BE PUBLISHED
|
|
5J0I
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 2L6HC3_12 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0M
 
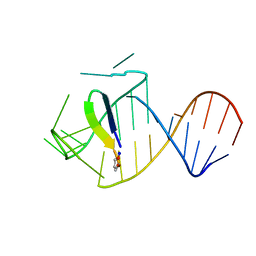 | | Ground state sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29-mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-28 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5MB1
 
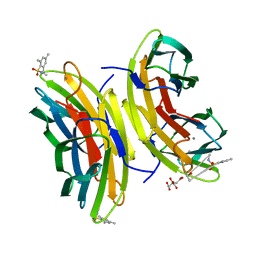 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PA14 IN COMPLEX WITH 2,4,6-Trimethylphenylsulfonamide-N-methyl-L-fucopyranoside | | Descriptor: | CALCIUM ION, CITRATE ANION, Fucose-binding lectin PA-IIL, ... | | Authors: | Sommer, R, Imberty, A, Titz, A, Varrot, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Glycomimetic, Orally Bioavailable LecB Inhibitors Block Biofilm Formation of Pseudomonas aeruginosa.
J. Am. Chem. Soc., 140, 2018
|
|
2N1C
 
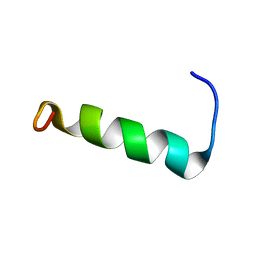 | | Structure of PvHCt, an antimicrobial peptide from shrimp litopenaeus vannamei | | Descriptor: | Hemocyanin subunit L2 | | Authors: | Petit, V.W, Rolland, J.L, Blond, A, Djediat, C, Peduzzi, J, Goulard, C, Bachere, E, Dupont, J, Destoumieux-Garzon, D, Rebuffat, S. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes.
Biochim.Biophys.Acta, 1860, 2015
|
|
5IXG
 
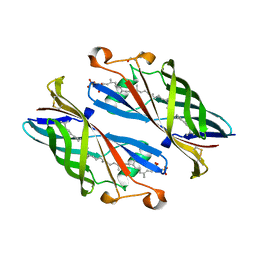 | | Crystal Structure of Burkholderia cenocepacia BcnB | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, YceI | | Authors: | Loutet, S.A, Murphy, M.E.P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Antibiotic Capture by Bacterial Lipocalins Uncovers an Extracellular Mechanism of Intrinsic Antibiotic Resistance.
MBio, 8, 2017
|
|
8DKU
 
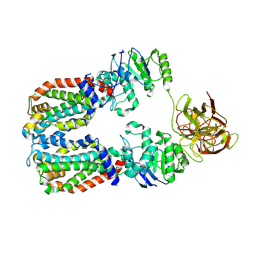 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, Transport permease protein | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
8DNC
 
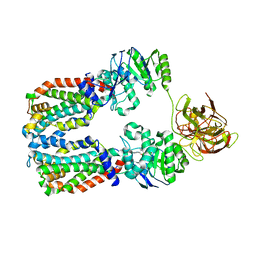 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen and ADP | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
6AWA
 
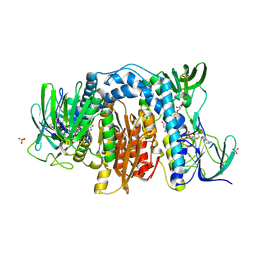 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6RL9
 
 | | Human Carbonic Anhydrase II in complex with 4-Aminobenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
2VCS
 
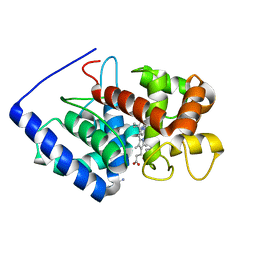 | | Structure of isoniazid (INH) bound to cytosolic soybean ascorbate peroxidase mutant H42A | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-09-26 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
4R9M
 
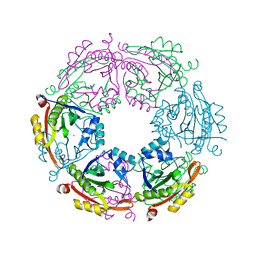 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
