5MPI
 
 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | Grainyhead-like protein 1 homolog | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
8P6M
 
 | |
5K0F
 
 | | Crystal Structure of COMT in complex with 5-[5-[1-(4-methoxyphenyl)ethyl]-1H-pyrazol-3-yl]-2,4-dimethyl-1,3-thiazole | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-{3-[(1R)-1-(4-methoxyphenyl)ethyl]-1H-pyrazol-5-yl}-2,4-dimethyl-1,3-thiazole, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
5MPN
 
 | | Crystal structure of CREBBP bromodomain complexed with FA26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethoxy-3-[3-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
8C39
 
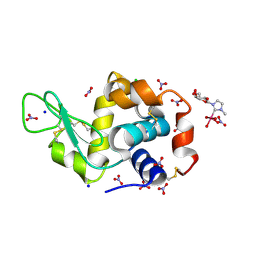 | | X-ray structure of HEWL upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | CHLORIDE ION, Lysozyme, NITRATE ION, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ruthenium(II)–Arene Complexes with Glycosylated NHC-Carbene Co-Ligands: Synthesis, Hydrolytic Behavior, and Binding to Biological Molecules
Organometallics, 42, 2023
|
|
5K0R
 
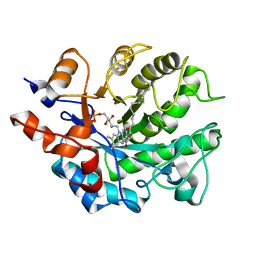 | | Crystal structure of reduced Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H:flavin oxidoreductase Sye4, Octadecane | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2016-05-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
6V7M
 
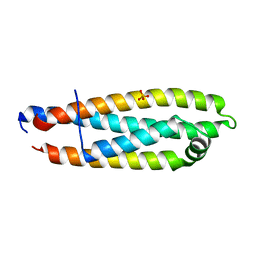 | |
4Y3Q
 
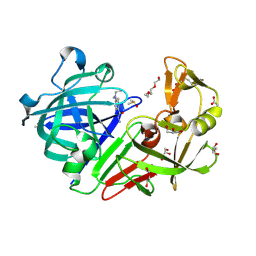 | |
5JVL
 
 | | C4-type pyruvate phospate dikinase: nucleotide binding domain with bound ATP analogue | | Descriptor: | 2'-Bromo-2'-deoxyadenosine 5'-[beta,gamma-imide]triphosphoric acid, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
6XC2
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
8ERD
 
 | | Cyclin-free CDK2 in complex with Cpd17 | | Descriptor: | (2-{[1-(methanesulfonyl)piperidin-4-yl]amino}quinazolin-7-yl)[(2S)-2-methylpyrrolidin-1-yl]methanone, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M, Oh, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Cyclin-free CDK2 in complex with Cpd17
To Be Published
|
|
6SOX
 
 | | Crystal structure of cAMP-dependent protein kinase A (CHO PKA) in complex with 4-carbamoylbenzoic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-aminocarbonylbenzoic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8TFZ
 
 | | tRNA 2'-phosphotransferase (Tpt1) from Pyrococcus horikoshii in complex with NAD | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8P9Y
 
 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
7NPE
 
 | | Vibrio cholerae ParA2-ADP | | Descriptor: | AAA family ATPase, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The cryo-EM structure of the bacterial type I segregation filament reveals ParA s conformational plasticity upon DNA binding
To Be Published
|
|
4Y51
 
 | | Endothiapepsin in complex with fragment 52 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-1-(4-bromophenyl)ethanone, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6VBE
 
 | |
5MT0
 
 | |
8PG0
 
 | | Human OATP1B3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHOLESTEROL, ... | | Authors: | Ciuta, A.-D, Nosol, K, Kowal, J, Mukherjee, S, Ramirez, A.S, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2023-06-17 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of human drug transporters OATP1B1 and OATP1B3.
Nat Commun, 14, 2023
|
|
8HEW
 
 | | Potato 14-3-3 St14f | | Descriptor: | 14-3-3 protein, StFDL1 peptide | | Authors: | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
6FA1
 
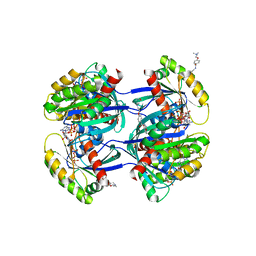 | | Antibody derived (Abd-4) small molecule binding to KRAS. | | Descriptor: | 2-[4-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methylcarbamoyl]phenoxy]ethyl-dimethyl-azanium, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
6SAA
 
 | | M-TRTX-Preg1a (Poecilotheria regalis) | | Descriptor: | U1-theraphotoxin-Pf3 | | Authors: | Meudal, H, Landon, C, Delmas, A.F. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-26 | | Method: | SOLUTION NMR | | Cite: | A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists.
J.Med.Chem., 63, 2020
|
|
6F91
 
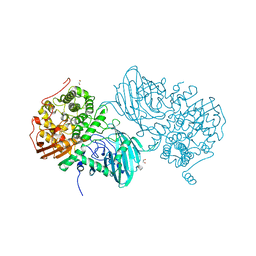 | | Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4Y6R
 
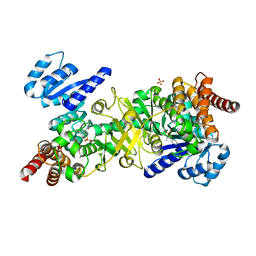 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, RC137, and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, CHLORIDE ION, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2015-02-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Bioactivity of beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase.
J.Med.Chem., 58, 2015
|
|
7N27
 
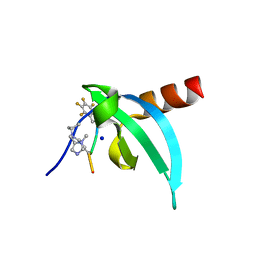 | | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261 | | Descriptor: | Isoform 2 of Chromodomain Y-like protein, SODIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Beldar, S, Dong, A, Loppnau, P, Min, J, Arrowsmith, C.H, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261
To Be Published
|
|
