6YNM
 
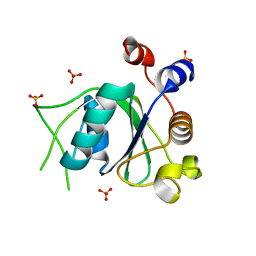 | | Crystal structure of YTHDC1 with compound DHU_DC1_096 | | Descriptor: | 1-[(2~{R},5~{S})-2-(3-chlorophenyl)-5-methyl-morpholin-4-yl]-3-methoxy-propan-2-ol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
5MHX
 
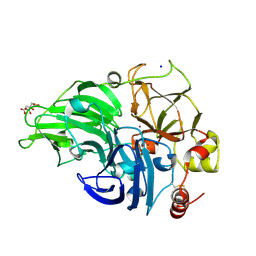 | | The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The sixth structure of the series with total exposition time 153 min. | | Descriptor: | CITRIC ACID, COPPER (II) ION, Laccase 2, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2016-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural study of the X-ray-induced enzymatic reduction of molecular oxygen to water by Steccherinum murashkinskyi laccase: insights into the reaction mechanism.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6FBW
 
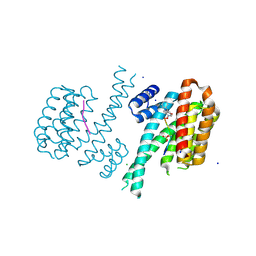 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-II with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
4XYF
 
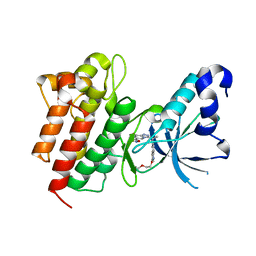 | | Crystal structure of c-Met in complex with (S)-5-(8-fluoro-3-(1-(3-(2-methoxyethoxy)quinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1S)-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-(2-methoxyethoxy)quinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
8UCU
 
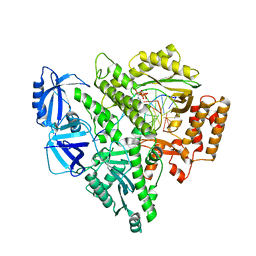 | | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5JOX
 
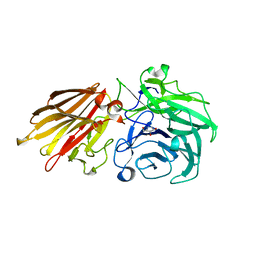 | | Bacteroides ovatus Xyloglucan PUL GH43A in complex with AraDNJ | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
4XZM
 
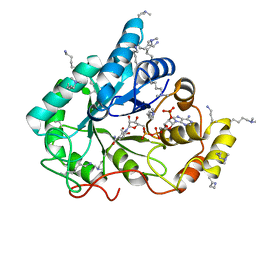 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
7SWT
 
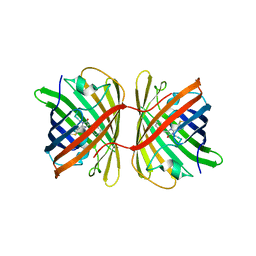 | | Crystal structure of the chromoprotein eforRED | | Descriptor: | Chromoprotein eforRED | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5MSA
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, CITRIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human carbonic anhydrase
isozyme XII with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide
To be published
|
|
5MQ6
 
 | | Polycyclic Ketone Monooxygenase from the Thermophilic Fungus Thermothelomyces thermophila | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Savino, S, Furst, M.J.L.J, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Polycyclic Ketone Monooxygenase from the Thermophilic Fungus Thermothelomyces thermophila: A Structurally Distinct Biocatalyst for Bulky Substrates.
J. Am. Chem. Soc., 139, 2017
|
|
6SKF
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-08-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
7AZR
 
 | |
8UCW
 
 | | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5MQT
 
 | | Crystal structure of dCK mutant C3S in complex with imatinib and UDP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Deoxycytidine kinase, ... | | Authors: | Saez-Ayala, M, Rebuffet, E, Hammam, K, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
7SWU
 
 | | Crystal structure of the chromoprotein spisPINK | | Descriptor: | Chromoprotein spisPINK | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7AZQ
 
 | |
5K18
 
 | | The NatB Acetyltransferase Complex Bound To bisubstrate inhibitor | | Descriptor: | Bisubstrate inhibitor, COENZYME A, N-terminal acetyltransferase B complex subunit NAT3, ... | | Authors: | Hong, H, Cai, Y, Zhang, S, Han, A. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB
Structure, 25, 2017
|
|
7TBC
 
 | | Crystal structure of Plasmepsin X from Plasmodium falciparum in complex with WM382 | | Descriptor: | (4R)-4-[(2E)-4,4-diethyl-2-imino-6-oxo-1,3-diazinan-1-yl]-N-[(4S)-2,2-dimethyl-3,4-dihydro-2H-1-benzopyran-4-yl]-3,4-dihydro-2H-1-benzopyran-6-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christensen, J.B, Hodder, A.N, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
6S9U
 
 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Ilumatobacter coccineus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
4XXA
 
 | | Crystal structure of a recombinant Vatairea macrocarpa seed lectin | | Descriptor: | CALCIUM ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Cunha, R.M.S, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2015-01-30 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a recombinant Vatairea macrocarpa seed lectin
Int.J.Biochem.Cell Biol., 2016
|
|
5JUD
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with uridine-diphosphate (UDP) - GpgS*UDP | | Descriptor: | Glucosyl-3-phosphoglycerate synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
7YWL
 
 | | Six DNA Helix Bundle nanopore - State 3 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
5MY0
 
 | | KS-MAT DI-DOMAIN OF MOUSE FAS WITH MALONYL-COA | | Descriptor: | COENZYME A, Fatty acid synthase, MALONYL-COENZYME A | | Authors: | Paithankar, K.S, Rittner, A, Huu, K.V, Grininger, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Characterization of the Polyspecific Transferase of Murine Type I Fatty Acid Synthase (FAS) and Implications for Polyketide Synthase (PKS) Engineering.
ACS Chem. Biol., 13, 2018
|
|
7YWI
 
 | | Six DNA duplex bundle nanopore - State 2 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWO
 
 | | Six DNA Helix Bundle nanopore - State 5 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
