2WBN
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 | | Descriptor: | TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-24 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
6VUW
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT368 | | Descriptor: | (7R)-7-methyl-2-({[(3R)-1-methylpiperidin-3-yl]methyl}sulfanyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-amine, GLYCEROL, N-acetyltransferase Eis, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
7AZT
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2I4T
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with Imm-A | | Descriptor: | 3,4-PYRROLIDINEDIOL,2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)-2S,3S,4R,5R, PHOSPHATE ION, Trichomonas vaginalis purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Schramm, V.L, Almo, S.C. | | Deposit date: | 2006-08-22 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues.
Biochemistry, 46, 2007
|
|
1JQU
 
 | |
4IBO
 
 | | Crystal structure of a putative gluconate dehydrogenase from agrobacterium tumefaciens (target EFI-506446) | | Descriptor: | CHLORIDE ION, Gluconate dehydrogenase, MAGNESIUM ION | | Authors: | Vetting, M.W, Bouvier, J.T, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative gluconate dehydrogenase from agrobacterium tumefaciens (target EFI-506446)
To be Published
|
|
7ZQ8
 
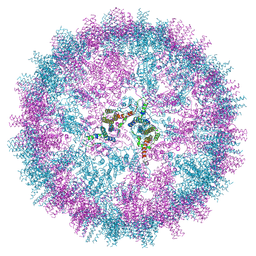 | | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=4 VLP) | | Descriptor: | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR | | Authors: | Kingston, N.J, Grehan, K, Snowden, J.S, Alzahrani, J, Ranson, N.A, Rowlands, D.J, Stonehouse, N.J. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | VelcroVax: a "Bolt-On" Vaccine Platform for Glycoprotein Display.
Msphere, 8, 2023
|
|
2VXY
 
 | | The structure of FTsZ from Bacillus subtilis at 1.7A resolution | | Descriptor: | CELL DIVISION PROTEIN FTSZ, CITRIC ACID, POTASSIUM ION | | Authors: | Barynin, V.V, Baker, P.J, Rice, D.W, Sedelnikova, S.E, Haydon, D.J, Stokes, N.R, Ure, R, Galbraith, G, Bennett, J.M, Brown, D.R, Heal, J.R, Sheridan, J.M, Aiwale, S.T, Chauhan, P.K, Srivastava, A, Taneja, A, Collins, I, Errington, J, Czaplewski, L.G. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Inhibitor of Ftsz with Potent and Selective Anti-Staphylococcal Activity.
Science, 321, 2008
|
|
6PNU
 
 | | Crystal structure of native DauA | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
7ANM
 
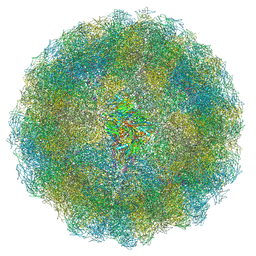 | | Nudaurelia capensis omega virus capsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
7ZQA
 
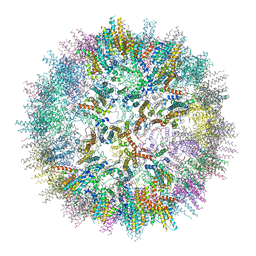 | | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=3* VLP) | | Descriptor: | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR | | Authors: | Kingston, N.J, Grehan, K, Snowden, J.S, Alzahrani, J, Ranson, N.A, Rowlands, D.J, Stonehouse, N.J. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | VelcroVax: a "Bolt-On" Vaccine Platform for Glycoprotein Display.
Msphere, 8, 2023
|
|
5KCE
 
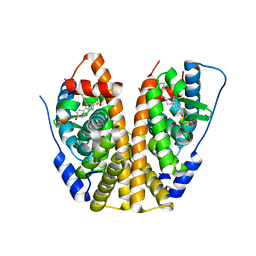 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl, 2-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(2-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-methyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCN
 
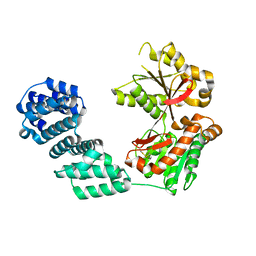 | |
6VX8
 
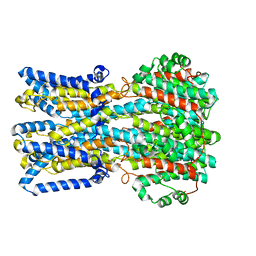 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
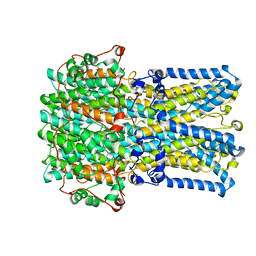 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2W03
 
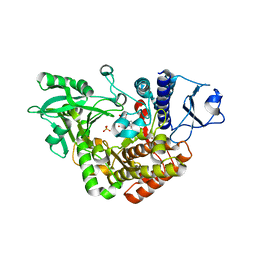 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with adenosine, sulfate and citrate from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE, CITRIC ACID, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
5KDN
 
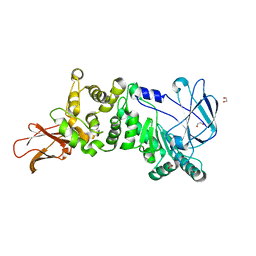 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, ZINC ION | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7ZVO
 
 | | Structure of CBM BT0996-C from Bacteroides thetaiotaomicron | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALANINE, BETA-MERCAPTOETHANOL, ... | | Authors: | Trovao, F, Pinheiro, B.A, Correia, V.G, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2022-05-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a Bacteroides thetaiotaomicron carbohydrate-binding module provides new insight into the recognition of complex pectic polysaccharides by the human microbiome.
J Struct Biol X, 7, 2023
|
|
5K3K
 
 | | Crystal structure of laccase from Thermus thermophilus HB27 (CuSO4, 20 min) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Diaz-Vilchis, A, Ruiz-Arellano, R.R, Rosas-Benitez, E, Rudino-Pinera, E. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Preserving metallic sites affected by radiation damage: the CuT2 case in Thermus thermophilus multicopper oxidase
To be Published
|
|
2W8V
 
 | | SPT with PLP, N100W | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
6W1B
 
 | |
3ZJI
 
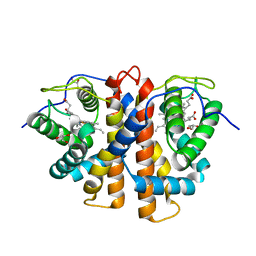 | | Tyr(61)B10Ala mutation of M.acetivorans protoglobin in complex with cyanide | | Descriptor: | CYANIDE ION, GLYCEROL, PROTOGLOBIN, ... | | Authors: | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
3X3C
 
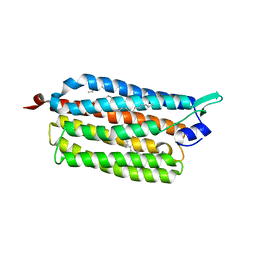 | | Crystal structure of the light-driven sodium pump KR2 in neutral state | | Descriptor: | OLEIC ACID, RETINAL, Sodium pumping rhodopsin | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
5K84
 
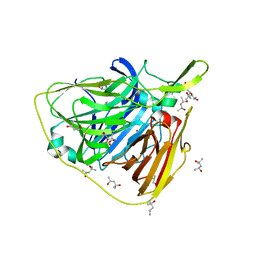 | | Crystal structure of laccase from Thermus thermophilus HB27 (sodium nitrate 10 min) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Laccase | | Authors: | Diaz-Vilchis, A, Ruiz-Arellano, R.R, Rosas-Benitez, E, Rudino-Pinera, E. | | Deposit date: | 2016-05-27 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Preserving metallic sites affected by radiation damage: the CuT2 case in Thermus thermophilus multicopper oxidase
To be Published
|
|
5D4W
 
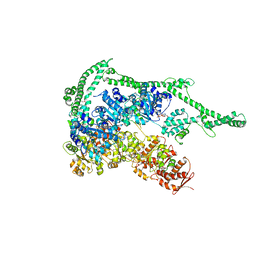 | | Crystal structure of Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative heat shock protein | | Authors: | Heuck, A, Schitter-Sollner, S, Clausen, T. | | Deposit date: | 2015-08-09 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the disaggregase activity and regulation of Hsp104.
Elife, 5, 2016
|
|
