8VDJ
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) as a covalent complex with EDP-235 | | Descriptor: | 3C-like proteinase nsp5, 4,6,7-trifluoro-N-{(2S)-1-[(3R,5'R)-5'-(iminomethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidin]-1'-yl]-4-methyl-1-oxopentan-2-yl}-N-methyl-1H-indole-2-carboxamide, THIOCYANATE ION | | Authors: | Cade, I.A, Rhodin, M.H.J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The small molecule inhibitor of SARS-CoV-2 3CLpro EDP-235 prevents viral replication and transmission in vivo.
Nat Commun, 15, 2024
|
|
7MFL
 
 | |
8E11
 
 | | Structure of mouse DNA polymerase Beta (PolB) mutant | | Descriptor: | ACETATE ION, DNA polymerase beta, MALONIC ACID | | Authors: | Sharma, N, Thompson, M.K, Prakash, A. | | Deposit date: | 2022-08-09 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pol beta /XRCC1 heterodimerization dictates DNA damage recognition and basal Pol beta protein levels without interfering with mouse viability or fertility.
DNA Repair (Amst), 123, 2023
|
|
8VHO
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
6UPS
 
 | |
8VHQ
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to dATP and ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
7M42
 
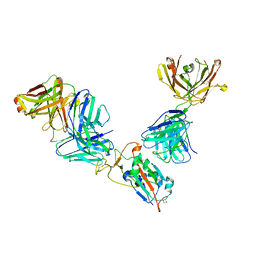 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of neutralizing antibodies REGN10985 and REGN10989 | | Descriptor: | REGN10985 antibody Fab fragment heavy chain, REGN10985 antibody Fab fragment light chain, REGN10989 antibody Fab fragment heavy chain, ... | | Authors: | Zhou, Y, Romero Hernandez, A, Saotome, K, Franklin, M.C. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies.
Cell, 184, 2021
|
|
8V09
 
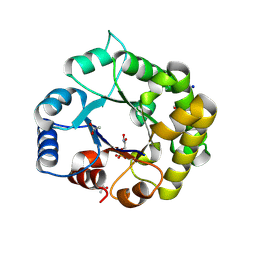 | | Crystal Structure of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by bayesian inference with PGH | | Descriptor: | ACETIC ACID, FORMIC ACID, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
7MFK
 
 | |
5MA5
 
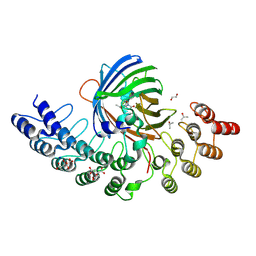 | | GFP-binding DARPin fusion gc_K11 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
6V1D
 
 | | Crystal structure of human trefoil factor 1 | | Descriptor: | Trefoil factor 1 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
7MLS
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Zahid, H, Buchholz, C.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
8VHR
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to dATP and GTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its apo form
To Be Published
|
|
5MFE
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with (RR)4 peptide | | Descriptor: | (RR)4, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
6P22
 
 | | Photorhabdus Virulence Cassette (PVC) PAAR repeat protein Pvc10 in complex with a T4 gp5 beta-helix fragment modified to mimic Pvc8, the central spike protein of PVC | | Descriptor: | 9-OCTADECENOIC ACID, CHIMERA OF CENTRAL SPIKE PROTEINS GP5 FROM PHAGE T4 AND PVC8 FROM PVC, MAGNESIUM ION, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2019-05-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Photorhabdus Virulence Cassette (PVC) PAAR repeat protein Pvc10 in complex with a T4 gp5 beta-helix fragment modified to mimic Pvc8, the central spike protein of PVC
To Be Published
|
|
5MDV
 
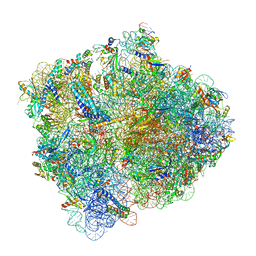 | | Structure of ArfA and RF2 bound to the 70S ribosome (accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MHL
 
 | |
8EBM
 
 | |
4QTB
 
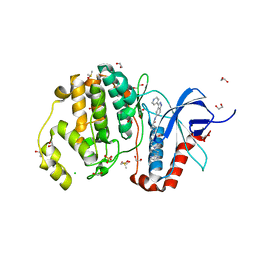 | | Structure of human ERK1 in complex with SCH772984 revealing a novel inhibitor-induced binding pocket | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Keates, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
8V2W
 
 | | Crystal Structure of the ancestral triosephosphate isomerase reconstruction of the last opisthokont common ancestor obtained by Bayesian inference | | Descriptor: | GLYCEROL, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8V0A
 
 | | Crystal Structure of the worst case of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8VHP
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to CDP and two molecules of ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8EBL
 
 | |
8VHN
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to two ATP molecules | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
