4UOH
 
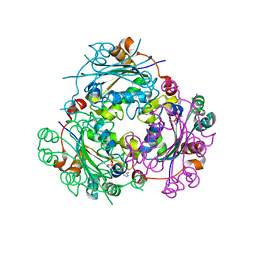 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Guevara-Hernandez, E, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4UIA
 
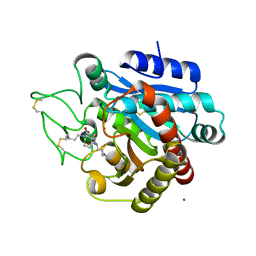 | | Crystal structure of 3a in complex with tafCPB | | Descriptor: | (2S)-6-azanyl-2-[[(2R)-3-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, CARBOXYPEPTIDASE B, ZINC ION | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4UHT
 
 | |
4USE
 
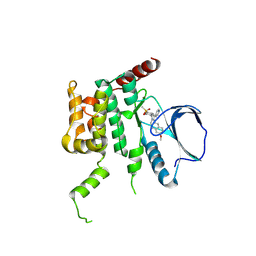 | | Human STK10 (LOK) with SB-633825 | | Descriptor: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
4UWA
 
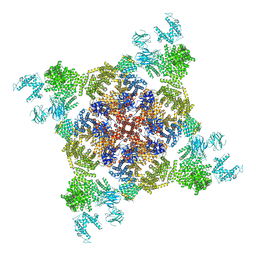 | |
4V48
 
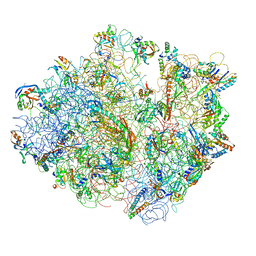 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4US5
 
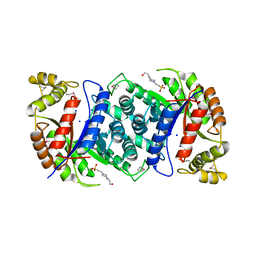 | | Crystal Structure of apo-MsnO8 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maier, S, Pflueger, T, Loesgen, S, Asmus, K, Broetz, E, Paululat, T, Zeeck, A, Andrade, S, Bechthold, A. | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Bioactivity of Mensacarcin and Epoxide Formation by Msno8.
Chembiochem, 15, 2014
|
|
4UWE
 
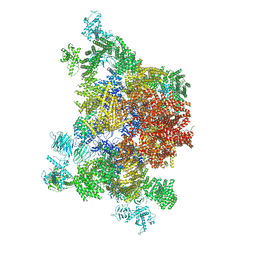 | |
4V4H
 
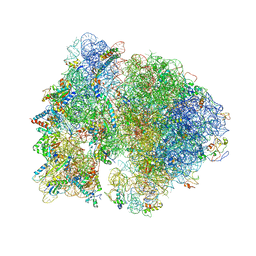 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with the antibiotic kasugamyin at 3.5A resolution. | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, ... | | Authors: | Schuwirth, B.S, Vila-Sanjurjo, A, Cate, J.H.D. | | Deposit date: | 2006-08-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structural analysis of kasugamycin inhibition of translation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4V7V
 
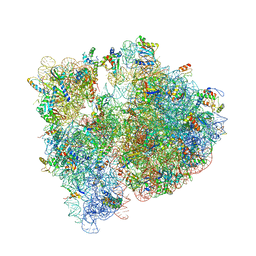 | | Crystal structure of the E. coli ribosome bound to clindamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.2891 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4USC
 
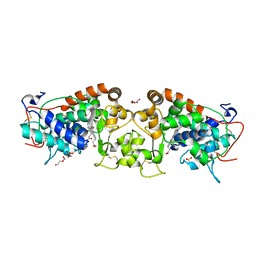 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
4UVM
 
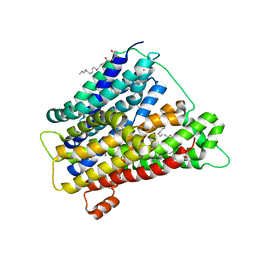 | | In meso crystal structure of the POT family transporter PepTSo | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLUTATHIONE UPTAKE TRANSPORTER | | Authors: | Lyons, J.A, Solcan, N, Caffrey, M, Newstead, S. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating Topology of the Proton-Coupled Oligopeptide Symporters.
Structure, 23, 2015
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
4V57
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with spectinomycin and neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Holton, J.M, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2007-07-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A steric block in translation caused by the antibiotic spectinomycin.
Acs Chem.Biol., 2, 2007
|
|
4W9A
 
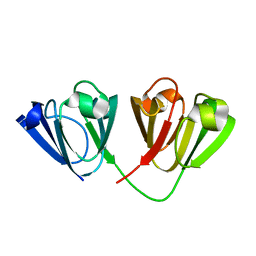 | | Crystal structure of Gamma-B Crystallin expressed in E. coli based on mRNA variant 2 | | Descriptor: | Gamma-crystallin B | | Authors: | Kudlinzki, D, Buhr, F, Linhard, V.L, Jha, S, Komar, A.A, Schwalbe, H. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Two synonymous gene variants encode proteins with identical sequence, but different folding conformations.
To Be Published
|
|
4UUU
 
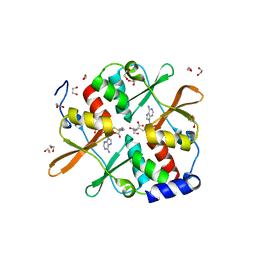 | | 1.7 A resolution structure of human cystathionine beta-synthase regulatory domain (del 516-525) in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, CYSTATHIONINE BETA-SYNTHASE, S-ADENOSYLMETHIONINE | | Authors: | Kopec, J, McCorvie, T.J, Fitzpatrick, F, Strain-Damerell, C, Froese, D.S, Tallant, C, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
4WBL
 
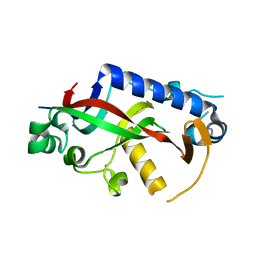 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4UV3
 
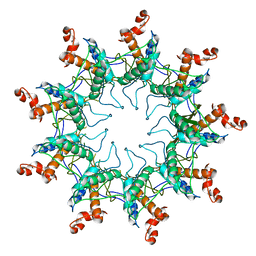 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4V3Q
 
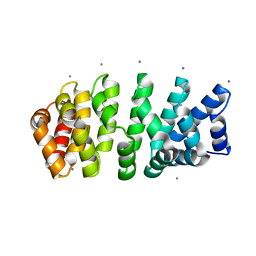 | | Designed armadillo repeat protein with 4 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | CALCIUM ION, GLYCEROL, YIII_M4_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4US7
 
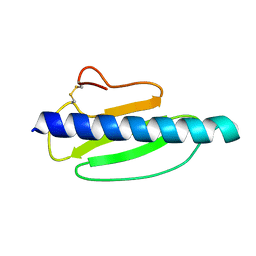 | | Sulfur SAD Phased Structure of a Type IV Pilus Protein from Shewanella oneidensis | | Descriptor: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | Authors: | Gorgel, M, Boeggild, A, Ulstrup, J.J, Mueller, U, Weiss, M, Nissen, P, Boesen, T. | | Deposit date: | 2014-07-03 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
4TSS
 
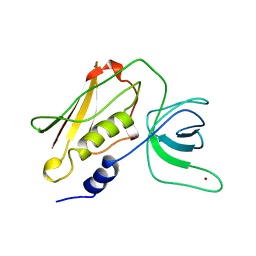 | | TOXIC SHOCK SYNDROME TOXIN-1: TETRAGONAL P4(1)2(1)2 CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1, ZINC ION | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
7CTS
 
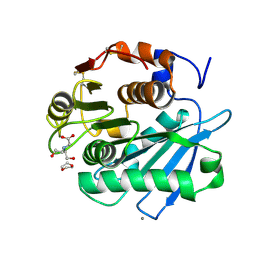 | | Open form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein, BICINE, ... | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
4V0Z
 
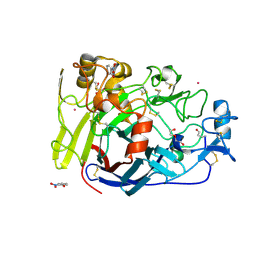 | |
4U0U
 
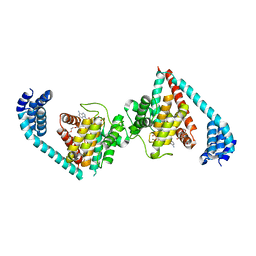 | | Wild type eukaryotic fic domain containing protein with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
7CVZ
 
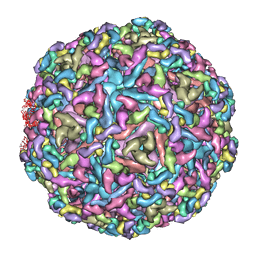 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
