4WQB
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - bisulfite soak | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEME C, IODIDE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.5013 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
2GEW
 
 | |
6E0S
 
 | | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library | | Descriptor: | GLYCEROL, MEM-A1, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Lau, C, Topp, E, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library
To Be Published
|
|
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
4WQ7
 
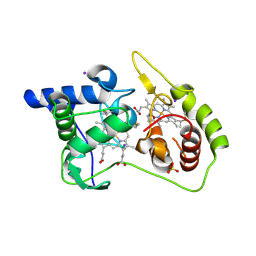 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - "as isolated" form | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WQE
 
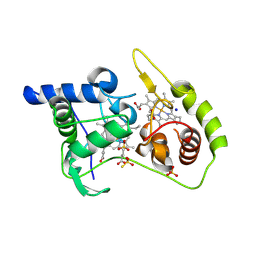 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - K208G mutant | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WFN
 
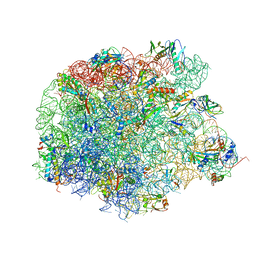 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 in complex with erythromycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-16 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
4WLK
 
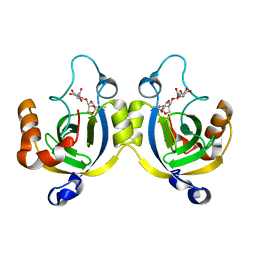 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with reaction product | | Descriptor: | N-[(1R,2S,3R,4R,5R)-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-6,8-dioxabicyclo[3.2.1]octan-4-yl]ethanamide, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WPG
 
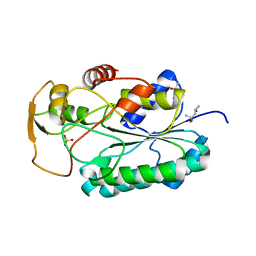 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
1HNO
 
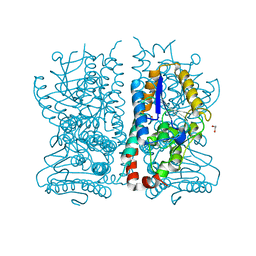 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1 | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
6DI9
 
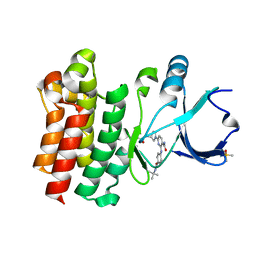 | | CRYSTAL STRUCTURE OF BTK IN COMPLEX WITH COVALENT INHIBITOR | | Descriptor: | 6-[(3S)-3-(acryloylamino)pyrrolidin-1-yl]-2-{[4-(tert-butylcarbamoyl)phenyl]amino}pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | GARDBERG, A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of potent, highly selective covalent irreversible BTK inhibitors from a fragment hit.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
2FY4
 
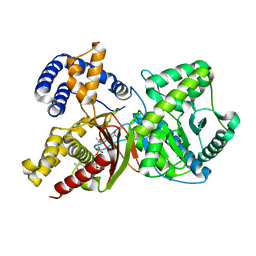 | |
3KOV
 
 | | Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte-specific enhancer factor 2A | | Authors: | Wu, Y, Dey, R, Han, A, Jayathilaka, N, Philips, M, Ye, J, Chen, L. | | Deposit date: | 2009-11-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the MADS-box/MEF2 Domain of MEF2A Bound to DNA and Its Implication for Myocardin Recruitment.
J.Mol.Biol., 397, 2010
|
|
4V0S
 
 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4F29
 
 | | Quisqualate bound to the ligand binding domain of GluA3i | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
6DIV
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-P5-2 (AJ-67) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-3-methyl-L-valyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methyl cyclopropyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diaz acyclopentadecine-14a(5H)-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
4UV9
 
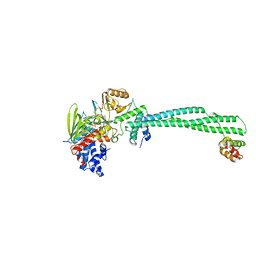 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
5KMY
 
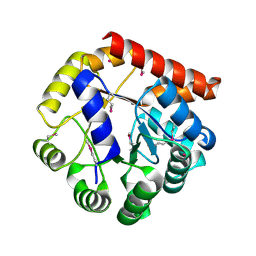 | | Crystal structure of tryptophan synthase subunit alpha from Legionella pneumophila str. Paris | | Descriptor: | Tryptophan synthase alpha chain | | Authors: | Nocek, B, Hatzos-Skintges, C, Endres, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-03 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Crystal structure of tryptophan synthase subunit alpha from Legionella pneumophila str. Paris
To Be Published
|
|
4UXL
 
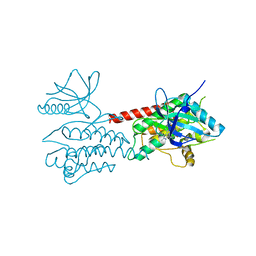 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4EYT
 
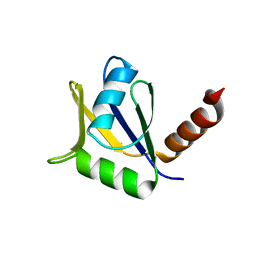 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 | | Descriptor: | SULFATE ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
2FOG
 
 | | Structure of porcine pancreatic elastase in 40% trifluoroethanol | | Descriptor: | CALCIUM ION, SULFATE ION, TRIFLUOROETHANOL, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
4UYK
 
 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
4F2H
 
 | |
4V0H
 
 | | Human metallo beta lactamase domain containing protein 1 (hMBLAC1) | | Descriptor: | FE (III) ION, GLYCEROL, METALLO-BETA-LACTAMASE DOMAIN-CONTAINING PROTEIN 1 1 | | Authors: | Pettinati, I, McDonough, M.A, Brem, J, Schofield, C.J. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Biosynthesis of histone messenger RNA employs a specific 3' end endonuclease.
Elife, 7, 2018
|
|
3KS8
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
