7VXX
 
 | | Zika virus NS2B/NS3 protease bZipro(C143S) in complex with 4-amino benzamidine | | Descriptor: | P-AMINO BENZAMIDINE, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Xiong, Y.C, Cheng, F, Zhang, J.Y, Su, H.X, Hu, H.C, Zou, Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of a novel inhibitor of the ZIKA virus NS2B/NS3 protease.
Bioorg.Chem., 128, 2022
|
|
4IVT
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(1H-indol-1-yl)acetamide, SULFATE ION | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
4IVS
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(6-cyano-1H-indol-1-yl)acetamide | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
6P7G
 
 | | The co-crystal structure of BRAF(V600E) with PHI1 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-[(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-[4-({[2-(morpholin-4-yl)ethyl]amino}methyl)-3-(trifluoromethyl)phenyl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Agianian, B, Gavathiotis, E. | | Deposit date: | 2019-06-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inhibitors of BRAF dimers using an allosteric site.
Nat Commun, 11, 2020
|
|
6P3D
 
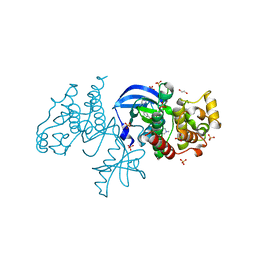 | | The co-crystal structure of BRAF(V600E) with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, AMMONIUM ION, ... | | Authors: | Agianian, B, Gavathiotis, E. | | Deposit date: | 2019-05-23 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Inhibitors of BRAF dimers using an allosteric site.
Nat Commun, 11, 2020
|
|
8DZH
 
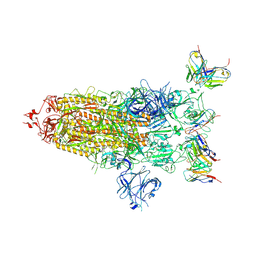 | |
8DZI
 
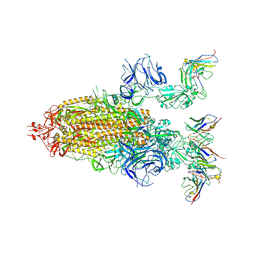 | |
4NGA
 
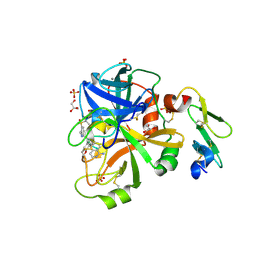 | |
4NG9
 
 | |
5OD6
 
 | |
5ODG
 
 | | Crystal structure of Smad3-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 3, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
6YPI
 
 | | Structure of the engineered metallo-Diels-Alderase DA7 W16G,K58Q,L77R,T78R | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BENZOIC ACID, DA7 W16G,K58Q,L77R,T78R, ... | | Authors: | Basler, S, Mori, T, Hilvert, D. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold.
Nat.Chem., 13, 2021
|
|
6UIP
 
 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
6EBA
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Unoccupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-08-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E8J
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Occupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E9C
 
 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
7BWW
 
 | | Structure of the engineered metallo-Diels-Alderase DA7 W16S | | Descriptor: | ACETIC ACID, BENZOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basler, S, Mori, T, Hilvert, D. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold.
Nat.Chem., 13, 2021
|
|
8DL5
 
 | |
8EQW
 
 | | Crystal structure of Fub7 | | Descriptor: | Sulfhydrylase FUB7 | | Authors: | Hai, Y. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular and Structural Basis for C gamma-C Bond Formation by PLP-Dependent Enzyme Fub7.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8ERJ
 
 | | Crystal structure of Fub7 in complex with E-2-aminocrotonate | | Descriptor: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]but-3-enoic acid, Sulfhydrylase FUB7 | | Authors: | Hai, Y. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Molecular and Structural Basis for C gamma-C Bond Formation by PLP-Dependent Enzyme Fub7.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8ERB
 
 | | Crystal structure of Fub7 in complex with vinylglycine ketimine | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, Sulfhydrylase FUB7 | | Authors: | Hai, Y. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular and Structural Basis for C gamma-C Bond Formation by PLP-Dependent Enzyme Fub7.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8W4Q
 
 | | Crystal structure of PDE4D complexed with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4R
 
 | | Crystal structure of PDE4D complexed with CVT-313 | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4S
 
 | | Crystal structure of PDE5A in complex with CVT-313 | | Descriptor: | 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4T
 
 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | 2-[bis(2-hydroxyethyl)amino]-6-[(4-methoxyphenyl)methylamino]-9-propan-2-yl-7~{H}-purin-8-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
