8D7X
 
 | |
8SAI
 
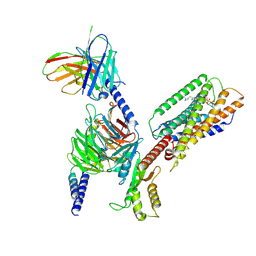 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5FFO
 
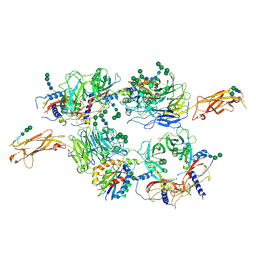 | | Integrin alpha V beta 6 in complex with pro-TGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Zhao, B, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
5FFG
 
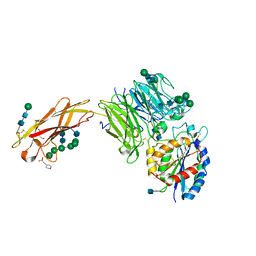 | | Crystal structure of integrin alpha V beta 6 head | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
5BPP
 
 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor 4AZ | | Descriptor: | 2-(4-butoxyphenyl)-N-hydroxyacetamide, ACETATE ION, Leukotriene A-4 hydrolase, ... | | Authors: | Huang, J, Dong, N.N, Xiao, Q, Ou, P.Y, Wu, D, Lu, W.Q. | | Deposit date: | 2015-05-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Bufexamac ameliorates LPS-induced acute lung injury in mice by targeting LTA4H
Sci Rep, 6, 2016
|
|
5H3R
 
 | | Crystal Structure of mutant MarR C80S from E.coli complexed with operator DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*CP*TP*TP*GP*CP*CP*TP*GP*GP*GP*CP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*TP*AP*TP*TP*GP*CP*CP*CP*AP*GP*GP*CP*AP*AP*GP*TP*AP*T)-3'), Multiple antibiotic resistance protein MarR | | Authors: | Zhu, R, Lou, H, Hao, Z. | | Deposit date: | 2016-10-26 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of the DNA-binding mechanism underlying the copper(II)-sensing MarR transcriptional regulator.
J. Biol. Inorg. Chem., 22, 2017
|
|
3HIZ
 
 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HHM
 
 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IEJ
 
 | | Pyrazole-based Cathepsin S Inhibitors with Arylalkynes as P1 Binding Elements | | Descriptor: | 2-[3-{4-chloro-3-[(4-chlorophenyl)ethynyl]phenyl}-1-(3-morpholin-4-ylpropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]-2-oxoethanol, Cathepsin S | | Authors: | Bembenek, S. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pyrazole-based cathepsin S inhibitors with arylalkynes as P1 binding elements.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6TO9
 
 | |
4JBA
 
 | |
6TO5
 
 | |
6TOC
 
 | |
3DBE
 
 | |
3DBD
 
 | |
3ML9
 
 | |
3ML8
 
 | |
5CD3
 
 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | Descriptor: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
3VOD
 
 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
8HK2
 
 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK3
 
 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK5
 
 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
3NGB
 
 | |
5TWN
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA- DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA- DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
