8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | Descriptor: | RNA (135-MER) | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5H3O
 
 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
2F4O
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, PHQ-VAL-ALA-ASP-CF0, XP-C repair complementing complex 58 kDa protein, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
2HPL
 
 | | Crystal structure of the mouse p97/PNGase complex | | Descriptor: | C-terminal of mouse p97/VCP, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HPJ
 
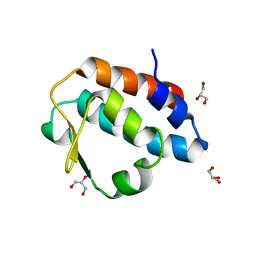 | | Crystal structure of the PUB domain of mouse PNGase | | Descriptor: | GLYCEROL, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5U6I
 
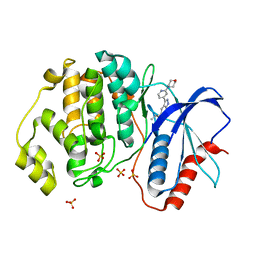 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
5TJA
 
 | | I-II linker of TRPML1 channel at pH 6 | | Descriptor: | Mucolipin-1 | | Authors: | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | Deposit date: | 2016-10-04 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TJC
 
 | | I-II linker of TRPML1 channel at pH 7.5 | | Descriptor: | Mucolipin-1 | | Authors: | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | Deposit date: | 2016-10-04 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7D5I
 
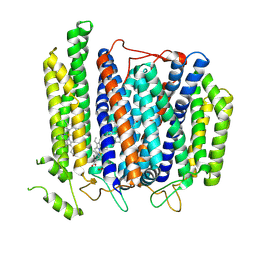 | | Structure of Mycobacterium smegmatis bd complex in the apo-form. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome D ubiquinol oxidase subunit 1, HEME B/C, ... | | Authors: | Wang, W, Gong, H, Gao, Y, Zhou, X, Rao, Z. | | Deposit date: | 2020-09-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Nat Commun, 12, 2021
|
|
8W4C
 
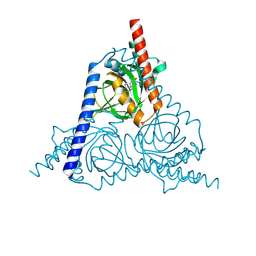 | | The sigma-1 receptor from Xenopus laevis in complex with progesterone by soaking | | Descriptor: | PROGESTERONE, Sigma non-opioid intracellular receptor 1 | | Authors: | Xiao, Y, Fu, C, Sun, Z, Zhou, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | Insight into binding of endogenous neurosteroid ligands to the sigma-1 receptor.
Nat Commun, 15, 2024
|
|
8W4B
 
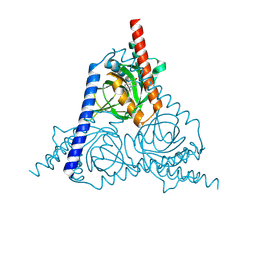 | | The sigma-1 receptor from Xenopus laevis in complex with progesterone by co-crystallization | | Descriptor: | PROGESTERONE, Sigma non-opioid intracellular receptor 1 | | Authors: | Xiao, Y, Fu, C, Sun, Z, Zhou, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Insight into binding of endogenous neurosteroid ligands to the sigma-1 receptor.
Nat Commun, 15, 2024
|
|
8WUE
 
 | |
8W4E
 
 | |
8W4D
 
 | |
8WWB
 
 | |
8YBB
 
 | |
8XIU
 
 | | Cryo-EM structure of a frog VMAT2 in an apo conformation | | Descriptor: | XlVMAT2 | | Authors: | Lyu, Y, Fu, C, Ma, H, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Engineering of a mammalian VMAT2 for cryo-EM analysis results in non-canonical protein folding.
Nat Commun, 15, 2024
|
|
8XIT
 
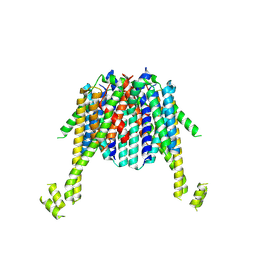 | | Cryo-EM structure of sheep VMAT2 dimer in an atypical fold | | Descriptor: | OaVMAT2-BRIL | | Authors: | Lyu, Y, Fu, C, Ma, H, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineering of a mammalian VMAT2 for cryo-EM analysis results in non-canonical protein folding.
Nat Commun, 15, 2024
|
|
7F5X
 
 | | GK domain of Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-L-GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5V
 
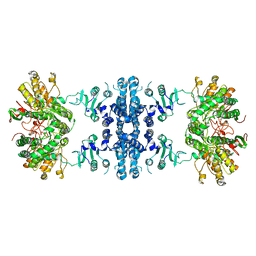 | | Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5U
 
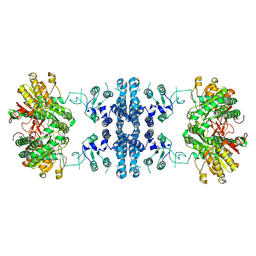 | |
