3IKA
 
 | |
4AZA
 
 | | Improved eIF4E binding peptides by phage display guided design. | | Descriptor: | EIF4G1_D5S PEPTIDE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Chew, W.Z, Quah, S.T, Verma, C.S, Liu, Y, Lane, D.P, Brown, C.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Improved Eif4E Binding Peptides by Phage Display Guided Design: Plasticity of Interacting Surfaces Yield Collective Effects.
Plos One, 7, 2012
|
|
8CYI
 
 | | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, N-[(2-aminoquinolin-7-yl)methyl]-9-(2-hydroxyethyl)-2,3,4,9-tetrahydro-1H-carbazole-6-carboxamide, ... | | Authors: | Yadav, G.P, Wei, Z, Xiaozhi, Y, Chenglong, L, Jiang, Q. | | Deposit date: | 2022-05-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Commun Biol, 5, 2022
|
|
6PMJ
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
2XKY
 
 | | Single particle analysis of Kir2.1NC_4 in negative stain | | Descriptor: | INWARD RECTIFIER POTASSIUM CHANNEL 2 | | Authors: | Fomina, S, Howard, T.D, Sleator, O.K, Golovanova, M, O'Ryan, L, Leyland, M.L, Grossmann, J.G, Collins, R.F, Prince, S.M. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17.200001 Å), SOLUTION SCATTERING | | Cite: | Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Biochim.Biophys.Acta, 1808, 2011
|
|
7S4E
 
 | |
3SUZ
 
 | | Crystal structure of Rat Mint2 PPC | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2 | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
5XLL
 
 | | Dimer form of M. tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
5XLM
 
 | | Monomer form of M.tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
3SV1
 
 | | Crystal structure of APP peptide bound rat Mint2 PARM | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2, Amyloid beta A4 protein | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
6IG5
 
 | |
6IGA
 
 | |
3LXF
 
 | | Crystal Structure of [2Fe-2S] Ferredoxin Arx from Novosphingobium aromaticivorans | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LXI
 
 | | Crystal Structure of Camphor-Bound CYP101D1 | | Descriptor: | CAMPHOR, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LXH
 
 | | Crystal Structure of Cytochrome P450 CYP101D1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LXD
 
 | | Crystal Structure of Ferredoxin Reductase ArR from Novosphingobium aromaticivorans | | Descriptor: | FAD-dependent pyridine nucleotide-disulphide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3E3R
 
 | |
3NV5
 
 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
3NV6
 
 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
1RTY
 
 | | Crystal Structure of Bacillus subtilis YvqK, a putative ATP-binding Cobalamin Adenosyltransferase, The North East Structural Genomics Target SR128 | | Descriptor: | PHOSPHATE ION, yvqk protein | | Authors: | Forouhar, F, Lee, I, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
5JJA
 
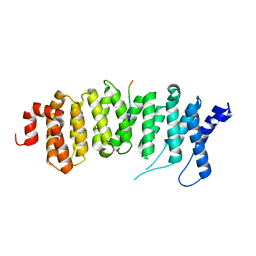 | | Crystal structure of a PP2A B56gamma/BubR1 complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Protein Cell, 7, 2016
|
|
1SQS
 
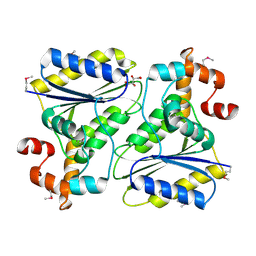 | | X-Ray Crystal Structure Protein SP1951 of Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR27. | | Descriptor: | L(+)-TARTARIC ACID, conserved hypothetical protein | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM, 8, 2007
|
|
1TM0
 
 | | Crystal Structure of the putative proline racemase from Brucella melitensis, Northeast Structural Genomics Target LR31 | | Descriptor: | PROLINE RACEMASE | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Ho, C.K, Ma, L.-C, Cooper, B, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-10 | | Release date: | 2004-06-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1ZBP
 
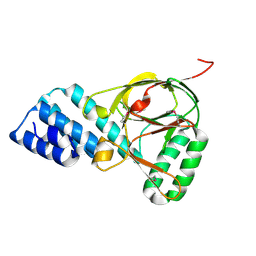 | | X-Ray Crystal Structure of Protein VPA1032 from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR44 | | Descriptor: | hypothetical protein VPA1032 | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Ciao, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
