2IHM
 
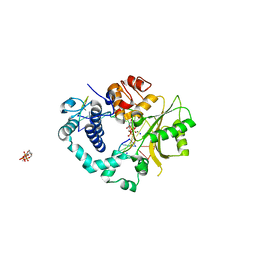 | | Polymerase mu in ternary complex with gapped 11mer DNA duplex and bound incoming nucleotide | | 分子名称: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', 5'-D(*CP*GP*GP*CP*AP*AP*TP*AP*CP*TP*G)-3', ... | | 著者 | Moon, A.F, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2006-09-26 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insight into the substrate specificity of DNA Polymerase mu.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2ID5
 
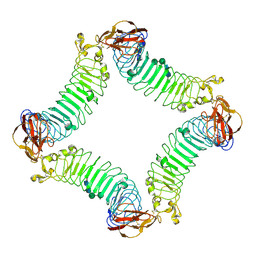 | | Crystal Structure of the Lingo-1 Ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | 著者 | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | 登録日 | 2006-09-14 | | 公開日 | 2006-09-26 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.698 Å) | | 主引用文献 | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
7UBU
 
 | |
7L4C
 
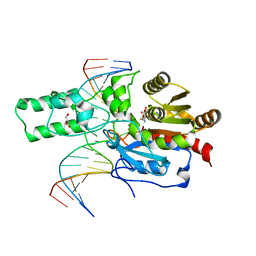 | | Crystal structure of the DRM2-CTT DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-18 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
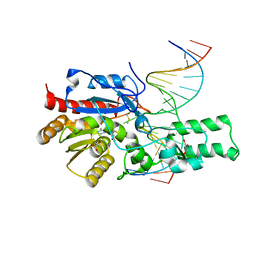 | | Crystal structure of the DRM2-CCG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
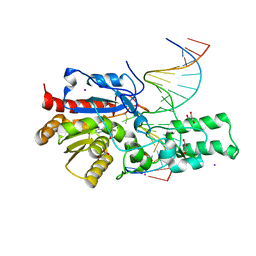 | | Crystal structure of the DRM2-CTG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
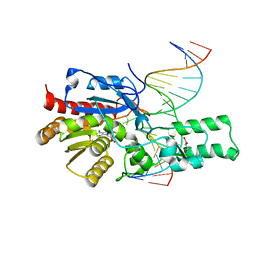 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.247 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
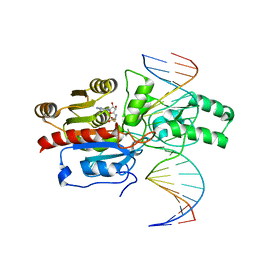 | | Crystal structure of the DRM2-CCT DNA complex | | 分子名称: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
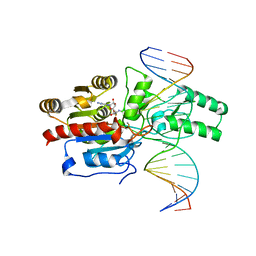 | | Crystal structure of the DRM2-CAT DNA complex | | 分子名称: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
5Z8N
 
 | |
5Z8L
 
 | |
8T1U
 
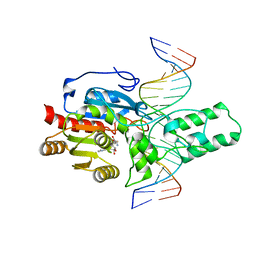 | | Crystal structure of the DRM2-CTA DNA complex | | 分子名称: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Chen, J, Lu, J, Song, J. | | 登録日 | 2023-06-03 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
3GAD
 
 | | Structure of apomif | | 分子名称: | ACETIC ACID, Macrophage migration inhibitory factor-like protein, SULFATE ION | | 著者 | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | 登録日 | 2009-02-17 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
3GAC
 
 | | Structure of mif with HPP | | 分子名称: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, Macrophage migration inhibitory factor-like protein, ... | | 著者 | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | 登録日 | 2009-02-17 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
6KN1
 
 | |
6KN0
 
 | |
6KMT
 
 | | P32 of caspase-11 mutant C254A | | 分子名称: | Caspase-4 | | 著者 | Ding, J, Sun, Q. | | 登録日 | 2019-08-01 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KMZ
 
 | |
6KMU
 
 | |
6KMV
 
 | |
4NJ5
 
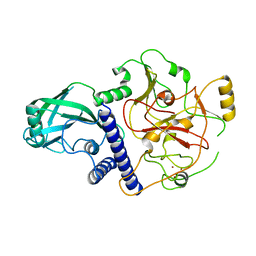 | | Crystal structure of SUVH9 | | 分子名称: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | 著者 | Du, J, Patel, D.J. | | 登録日 | 2013-11-08 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
5ZNP
 
 | |
5ZNR
 
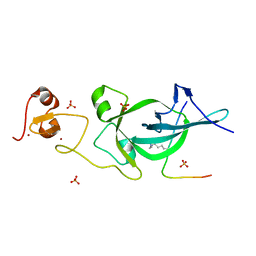 | |
4E3C
 
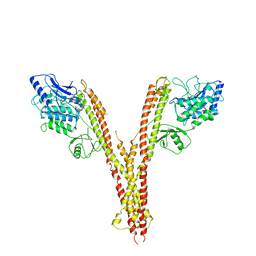 | | X-ray crystal structure of human IKK2 in an active conformation | | 分子名称: | Inhibitor of nuclear factor kappa-B kinase subunit beta | | 著者 | Polley, S, Huang, D.B, Hauenstein, A.V, Ghosh, G, Huxford, T. | | 登録日 | 2012-03-09 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.98 Å) | | 主引用文献 | X-ray crystal structure of human IKK2 in an active conformation
Plos.Biol., 2013
|
|
