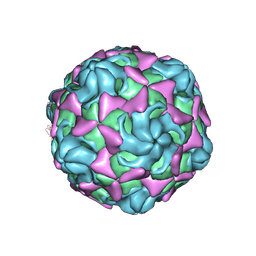
6LA6
 
 | |
6LIY
 
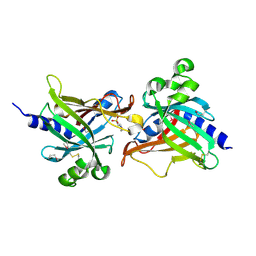 | | SeMet CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
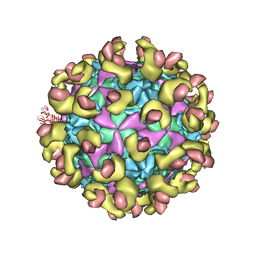
6LAP
 
 | | Cryo-EM structure of echovirus 11 A-particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
5XZ3
 
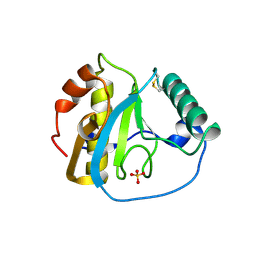 | | The X-ray structure of Apis mellifera PGRP-SA | | Descriptor: | Peptidoglycan-recognition protein, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae).
Protein Sci., 27, 2018
|
|
8HPU
 
 | |
8HPF
 
 | |
8HQ7
 
 | |
8HPV
 
 | |
8HP9
 
 | |
8HPQ
 
 | |
6A0Q
 
 | | The crystal structure of Lpg2622_E64 complex | | Descriptor: | Lpg2622, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
6A0N
 
 | | The crystal structure of apo-Lpg2622 | | Descriptor: | Lpg2622 | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
7YHW
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.12.1 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8DVG
 
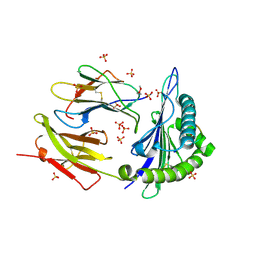 | | Structure of KRAS WT(7-16)-HLA-A*03:01 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Wright, K.M, Miller, M, Gabelli, S.B. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
2M6X
 
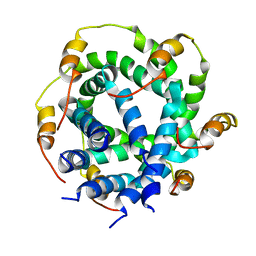 | |
2N2N
 
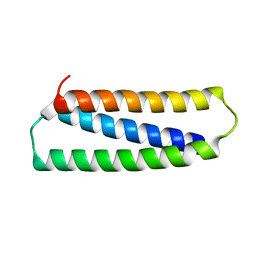 | |
8GD0
 
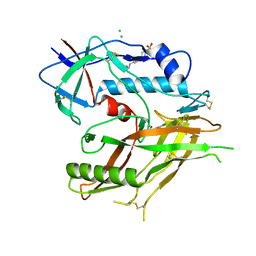 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with TFH-I-070-A6 | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-(4-methylpiperazine-1-carbonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD1
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with ZXC-I-092 | | Descriptor: | (3S,5S)-5-(aminomethyl)-N-(4-chloro-3-fluorophenyl)-1-[(3R,5S)-3,4,5-trimethylpiperazine-1-carbonyl]piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GJT
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with TFH-I-116-D1 | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-[(3R,5S)-3,4,5-trimethylpiperazine-1-carbonyl]piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GCZ
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with ZXC-I-090 | | Descriptor: | (3S,5S)-5-(aminomethyl)-N-(4-chloro-3-fluorophenyl)-1-(4-methylpiperazine-1-carbonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nguyen, D.N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD5
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD3
 
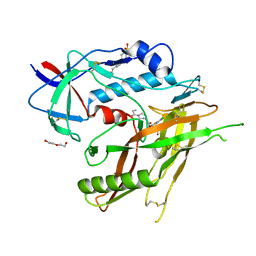 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-101 | | Descriptor: | (3S,5R)-N-(4-chloro-3-fluorophenyl)-1-(4-glycylpiperazine-1-carbonyl)-5-(hydroxymethyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
7YJ3
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
2N31
 
 | |
8H92
 
 | |
