4OV4
 
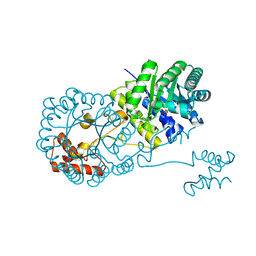 | | Isopropylmalate synthase binding with ketoisovalerate | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
6WGN
 
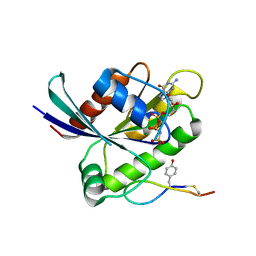 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | Descriptor: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
3UXY
 
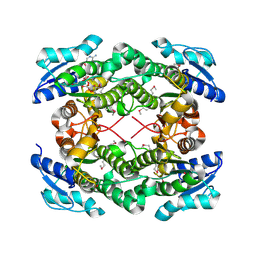 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
3UOE
 
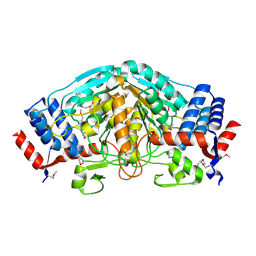 | | The crystal structure of dehydrogenase from Sinorhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The crystal structure of dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3V76
 
 | | The crystal structure of a flavoprotein from Sinorhizobium meliloti | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavoprotein | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of a flavoprotein from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
1MD2
 
 | | CHOLERA TOXIN B-PENTAMER WITH DECAVALENT LIGAND BMSC-0013 | | Descriptor: | 3-ETHYLAMINO-4-METHYLAMINO-CYCLOBUTANE-1,2-DIONE, CHOLERA TOXIN B SUBUNIT, CYANIDE ION, ... | | Authors: | Zhang, Z, Merritt, E.A, Ahn, M, Roach, C, Hol, W.G.J, Fan, E. | | Deposit date: | 2002-08-06 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Solution and crystallographic studies of branched multivalent ligands that inhibit the receptor-binding of cholera toxin.
J.Am.Chem.Soc., 124, 2002
|
|
6CO7
 
 | | Structure of the nvTRPM2 channel in complex with Ca2+ | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, Z, Toth, B, Szollosi, A, Chen, J, Csanady, L. | | Deposit date: | 2018-03-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure of a TRPM2 channel in complex with Ca2+explains unique gating regulation.
Elife, 7, 2018
|
|
1JW6
 
 | | Crystal Structure of the Complex of Concanavalin A and Hexapeptide | | Descriptor: | CALCIUM ION, Concanavalin A, ISOPROPYL ALCOHOL, ... | | Authors: | Zhang, Z, Qian, M, Huang, Q, Jia, Y, Tang, Y. | | Deposit date: | 2001-09-02 | | Release date: | 2001-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the complex of concanavalin A and hexapeptide.
J.Protein Chem., 20, 2001
|
|
8SK6
 
 | | human liver mitochondrial Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | | Descriptor: | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-18 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
1M4Z
 
 | | Crystal structure of the N-terminal BAH domain of Orc1p | | Descriptor: | MANGANESE (II) ION, ORIGIN RECOGNITION COMPLEX SUBUNIT 1 | | Authors: | Zhang, Z, Hayashi, M.K, Merkel, O, Stillman, B, Xu, R.-M. | | Deposit date: | 2002-07-05 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing.
EMBO J., 21, 2002
|
|
1JJR
 
 | |
8BDP
 
 | |
3LIF
 
 | |
3LIE
 
 | |
3LIB
 
 | |
3LIC
 
 | |
3LID
 
 | |
3LI9
 
 | |
3LI8
 
 | |
3LNV
 
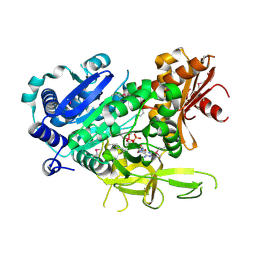 | | The crystal structure of fatty acyl-adenylate ligase from L. pneumophila in complex with acyl adenylate and pyrophosphate | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, PYROPHOSPHATE 2-, Saframycin Mx1 synthetase B | | Authors: | Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies of Fatty Acyl Adenylate Ligases from E. coli and L. pneumophila.
J.Mol.Biol., 406, 2011
|
|
3LIA
 
 | |
3LL3
 
 | |
7OQE
 
 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQB
 
 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
