7YBG
 
 | |
8Y7T
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with C2 | | Descriptor: | 3C-like proteinase nsp5, 6-(iminomethyl)-4-(2-pyridin-2-ylethyl)-2-[4-(trifluoromethyl)phenyl]-1,2,4-triazine-3,5-dione | | Authors: | Zeng, R, Deng, X.Y, Yang, Z.Y, Wang, K, Jiang, Y.Y, Lei, J. | | Deposit date: | 2024-02-05 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A deep learning model for structure-based bioactivity optimization and its application in the bioactivity optimization of a SARS-CoV-2 main protease inhibitor.
Eur.J.Med.Chem., 291, 2025
|
|
8Y7U
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with C5 | | Descriptor: | 2-(3-fluoro-4-(trifluoromethyl)phenyl)-6-(iminomethyl)-4-(2-oxo-2-(pyridin-2-yl)ethyl)-1,2,4-triazine-3,5(2H,4H)-dione, 3C-like proteinase nsp5 | | Authors: | Zeng, R, Deng, X.Y, Yang, Z.Y, Wang, K, Jiang, Y.Y, Lei, J. | | Deposit date: | 2024-02-05 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A deep learning model for structure-based bioactivity optimization and its application in the bioactivity optimization of a SARS-CoV-2 main protease inhibitor.
Eur.J.Med.Chem., 291, 2025
|
|
7C7P
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
8HHT
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Hit-1 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | | Authors: | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2022-11-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
8HHU
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with SY110 | | Descriptor: | (1~{R})-3,3-bis(fluoranyl)-~{N}-[(2~{R})-3-methoxy-1-oxidanylidene-1-[[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]amino]propan-2-yl]cyclohexane-1-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2022-11-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I30
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 32j | | Descriptor: | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Zeng, R, Huang, C, Xie, L.W, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and structure-activity relationship studies of novel alpha-ketoamide derivatives targeting the SARS-CoV-2 main protease.
Eur.J.Med.Chem., 259, 2023
|
|
7C8B
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Z-VAD(OMe)-FMK | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK
To Be Published
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
8JOP
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | Descriptor: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | Authors: | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | Deposit date: | 2023-06-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
7FAY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with (R)-1a | | Descriptor: | (2~{R})-~{N}-[(1~{R})-2-(~{tert}-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-~{N}-(4-~{tert}-butylphenyl)-2-oxidanyl-propanamide, 3C-like proteinase | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
7FAZ
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Y180 | | Descriptor: | (2~{R})-~{N}-dibenzofuran-3-yl-~{N}-[(1~{R})-2-[[(1~{S})-1-(4-fluorophenyl)ethyl]amino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]-2-oxidanyl-propanamide, 3C-like proteinase, SODIUM ION | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
7CX9
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1 | | Descriptor: | 3-iodanyl-1~{H}-indazole-7-carbaldehyde, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zeng, R, Liu, X.L, Qiao, J.X, Nan, J.S, Wang, Y.F, Li, Y.S, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1
To Be Published
|
|
8W8Z
 
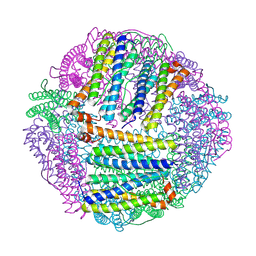 | | Escherichia coli ferritin mutant-M52H | | Descriptor: | Bacterioferritin | | Authors: | Zhao, G, Zeng, R. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli ferritin mutant M52H at 2.05 Angstroms resolution
To Be Published
|
|
8ZAI
 
 | |
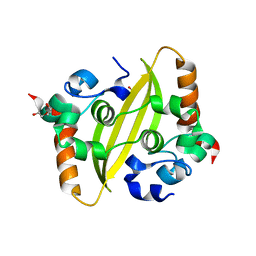
7C22
 
 | |
9IZB
 
 | |
6F7A
 
 | | Gloeobacter Ligand-gated Ion Channel (GLIC) closed state crystallized in an ultra-swollen lipidic mesophase | | Descriptor: | Proton-gated ion channel | | Authors: | Martiel, I, Zabara, A, Chong, J.Y.T, Stark, L, Cromer, B, Dummond, C.J, Mezzenga, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Design of ultra-swollen lipidic mesophases for the crystallization of membrane proteins with large extracellular domains.
Nat Commun, 9, 2018
|
|
8WTI
 
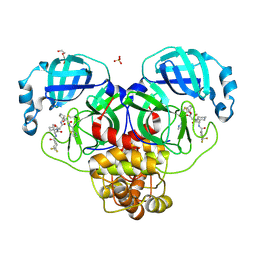 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
3QAH
 
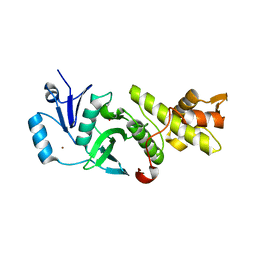 | | Crystal structure of apo-form human MOF catalytic domain | | Descriptor: | Probable histone acetyltransferase MYST1, ZINC ION | | Authors: | Sun, B, Tang, Q, Zhong, C, Ding, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274
Cell Res., 21, 2011
|
|
9J5S
 
 | | Crystal structure of human G3BP1 in complex with CHIKV nsP3 peptide | | Descriptor: | Polyprotein P1234, Ras GTPase-activating protein-binding protein 1 | | Authors: | Liu, Y.Z, Lei, J. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Cryo-EM reveals a double oligomeric ring scaffold of the CHIKV nsP3 peptide in complex with the NTF2L domain of host G3BP1.
Mbio, 16, 2025
|
|
9IVQ
 
 | | Cryo-EM structure of the CHIKV nsP3 peptide in complex with the NTF2L domain of G3BP1 (Conformation I) | | Descriptor: | Polyprotein P1234, Ras GTPase-activating protein-binding protein 1 | | Authors: | Wang, J, Liu, Y.Z, Lei, J, Wang, K.T. | | Deposit date: | 2024-07-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-EM reveals a double oligomeric ring scaffold of the CHIKV nsP3 peptide in complex with the NTF2L domain of host G3BP1.
Mbio, 16, 2025
|
|
9IVR
 
 | | Cryo-EM structure of the CHIKV nsP3 peptide in complex with the NTF2L domain of G3BP1 (Conformation II) | | Descriptor: | Polyprotein P1234, Ras GTPase-activating protein-binding protein 1 | | Authors: | Wang, J, Liu, Y.Z, Lei, J, Wang, K.T. | | Deposit date: | 2024-07-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM reveals a double oligomeric ring scaffold of the CHIKV nsP3 peptide in complex with the NTF2L domain of host G3BP1.
Mbio, 16, 2025
|
|
