8GOE
 
 | | Structure of hSLC19A1+5-MTHF | | Descriptor: | N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
1OZ1
 
 | | P38 MITOGEN-ACTIVATED KINASE IN COMPLEX WITH 4-AZAINDOLE INHIBITOR | | Descriptor: | 3-(4-FLUOROPHENYL)-2-PYRIDIN-4-YL-1H-PYRROLO[3,2-B]PYRIDIN-1-OL, Mitogen-activated protein kinase 14 | | Authors: | Lovejoy, B, Villasenor, A, Browner, M, Dunten, P. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of 4-azaindoles as inhibitors of p38 MAP kinase.
J.Med.Chem., 46, 2003
|
|
5ULA
 
 | |
2G46
 
 | |
1HZM
 
 | |
1M3G
 
 | |
6DUV
 
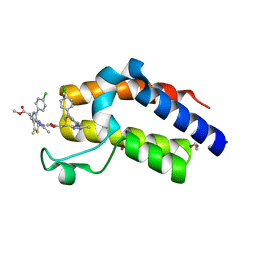 | | Crystal structure of the second bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Babault, N, Zhou, M.M. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Bound Water Molecules in Differential Recognition of Di-Acetylated Histone Peptides by the BRD4 Bromodomains
To be published
|
|
3DWH
 
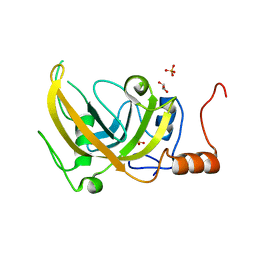 | |
8I60
 
 | |
7E1Q
 
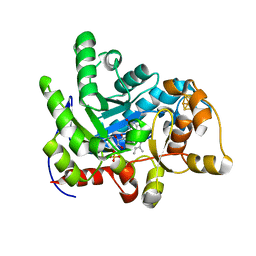 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori | | Descriptor: | 2-nitropropane dioxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1S
 
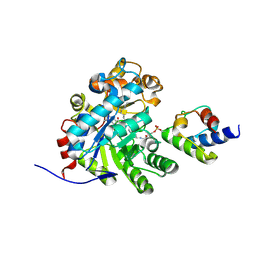 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori in complex with octanoyl-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1R
 
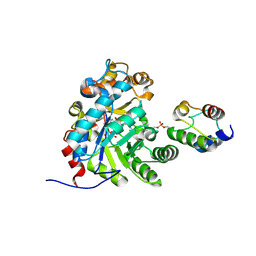 | | Crystal structure of Dehydrogenase/isomerase FabX from Helicobacter pylori in complex with holo-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
2FE9
 
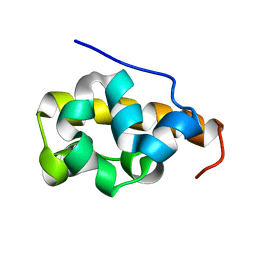 | |
4NUC
 
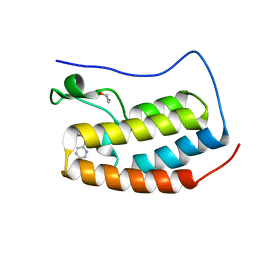 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS435 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(4-hydroxy-3,5-dimethylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
4NUE
 
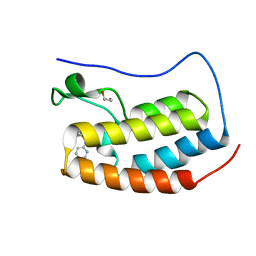 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS267 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(2-amino-4-hydroxy-3,5-dimethylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
4NUD
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS436 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
6LH8
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
4F3I
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Joshua, J, Zhou, M.-M, Plotnikov, A.N. | | Deposit date: | 2012-05-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Down-regulation of NF-{kappa}B Transcriptional Activity in HIV-associated Kidney Disease by BRD4 Inhibition.
J.Biol.Chem., 287, 2012
|
|
2LSP
 
 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
5ZGU
 
 | |
5ZGE
 
 | | Crystal structure of NDM-1 at pH5.5 (Bis-Tris) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGR
 
 | | Crystal structure of NDM-1 at pH7.3 (HEPES) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGV
 
 | |
5ZGY
 
 | |
