8FU7
 
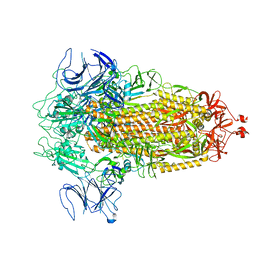 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU8
 
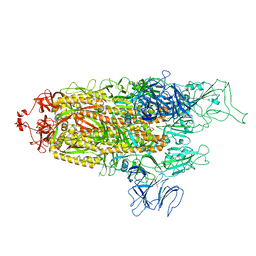 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU3
 
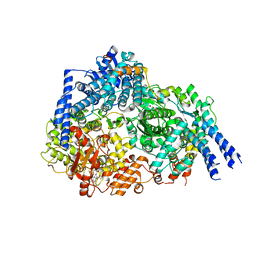 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
3J2V
 
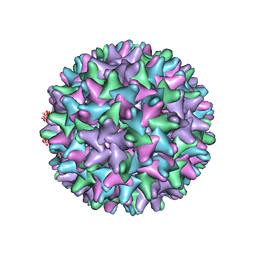 | | CryoEM structure of HBV core | | Descriptor: | PreC/core protein | | Authors: | Yu, X, Jin, L, Jih, J, Shih, C, Zhou, Z.H. | | Deposit date: | 2013-01-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein.
Plos One, 8, 2013
|
|
3J1R
 
 | | Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-terminal Type IV Pilin Helices | | Descriptor: | archaeal adhesion filament core | | Authors: | Yu, X, Goforth, C, Meyer, C, Rachel, R, Wirth, R, Schroeder, G.F, Egelman, E.H. | | Deposit date: | 2012-05-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-Terminal Type IV Pilin Helices.
J.Mol.Biol., 422, 2012
|
|
3TB0
 
 | |
5CAZ
 
 | | Crystallographic structure of apo human rotavirus K8 VP8* | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Outer capsid protein VP4, ... | | Authors: | Yu, X, Blanchard, H. | | Deposit date: | 2015-06-30 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substantial Receptor-induced Structural Rearrangement of Rotavirus VP8*: Potential Implications for Cross-Species Infection.
Chembiochem, 16, 2015
|
|
5CA6
 
 | |
5CB7
 
 | |
5F3O
 
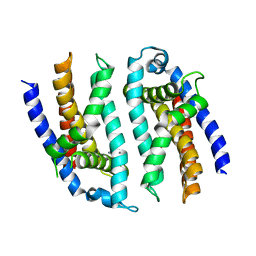 | |
5F3Q
 
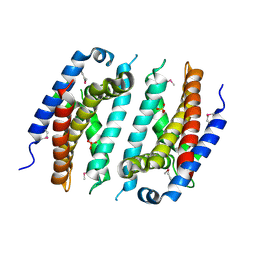 | |
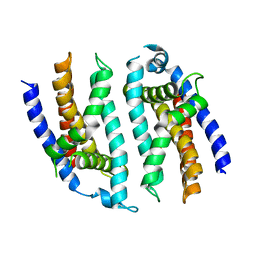
5F3P
 
 | |
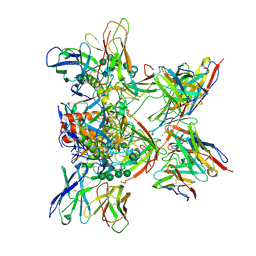
8DPL
 
 | |
4JZC
 
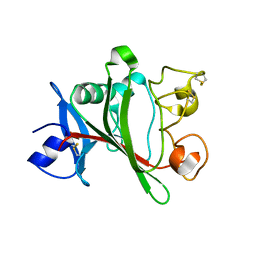 | | Angiopoietin-2 fibrinogen domain TAG mutant | | Descriptor: | Angiopoietin-2 | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-04-02 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0V
 
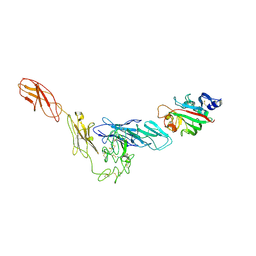 | | Structural basis for angiopoietin-1 mediated signaling initiation | | Descriptor: | Angiopoietin-1, TEK tyrosine kinase variant | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYO
 
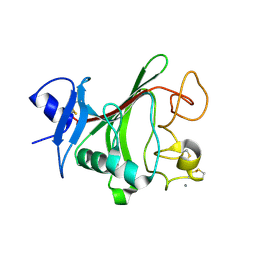 | | Structural basis for angiopoietin-1 mediated signaling initiation | | Descriptor: | Angiopoietin-1, CALCIUM ION | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
6MPB
 
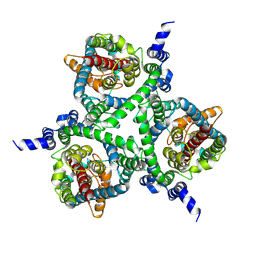 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
8A1S
 
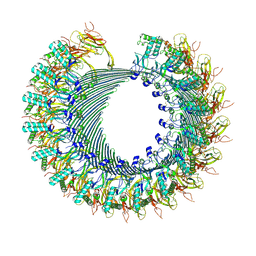 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
8A1D
 
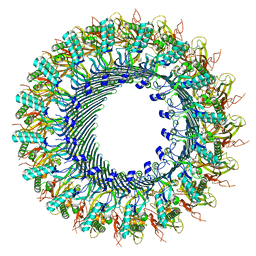 | | Structure of murine perforin-2 (Mpeg1) pore in ring form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
7CX0
 
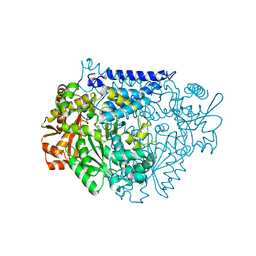 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa | | Descriptor: | CARBIDOPA, Decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa
to be published
|
|
7CWX
 
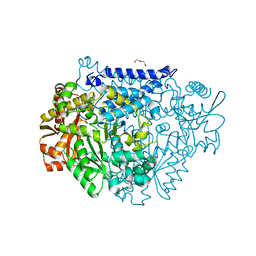 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Decarboxylase, GLYCEROL | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis
to be published
|
|
7CWY
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP | | Descriptor: | Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP
to be published
|
|
7CX1
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine | | Descriptor: | 4-[(2R)-2-(methylamino)propyl]phenol, Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine
to be published
|
|
7CWZ
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa | | Descriptor: | Decarboxylase, L-DOPAMINE, MAGNESIUM ION, ... | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa
to be published
|
|
