2PE2
 
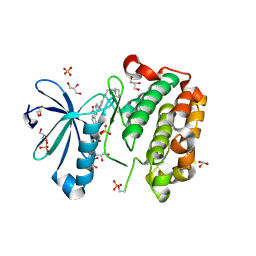 | |
7XRQ
 
 | |
6K1N
 
 | | PLP-bound form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
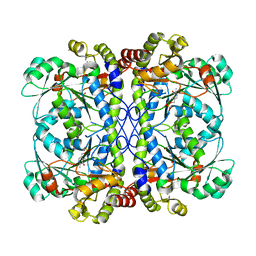
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6MUM
 
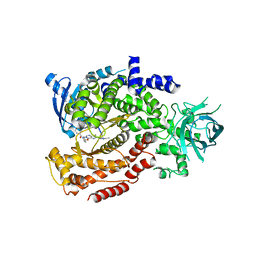 | | Murine PI3K delta kinsae domain - cpd 3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, [(3S)-3-{[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]oxy}pyrrolidin-1-yl](oxan-4-yl)methanone | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
6MUL
 
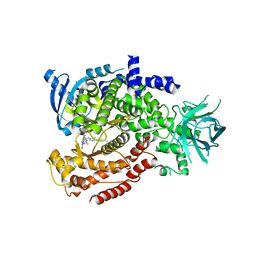 | | Murine PI3K delta kinsae domain - cpd 1 | | Descriptor: | 1-{1-[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]piperidin-4-yl}-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
7XE4
 
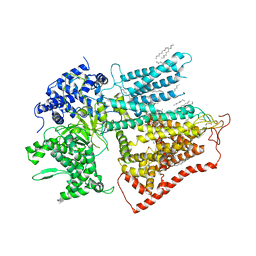 | | structure of a membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
7YUY
 
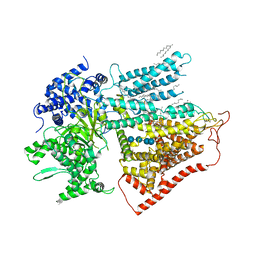 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
5WHT
 
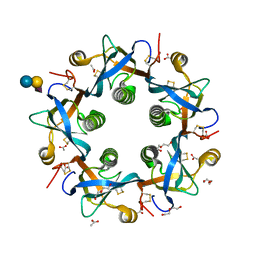 | | Crystal structure of 3'SL bound PltB | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Gao, X, Galan, J.E. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Evolution of host adaptation in the Salmonella typhoid toxin.
Nat Microbiol, 2, 2017
|
|
5WHU
 
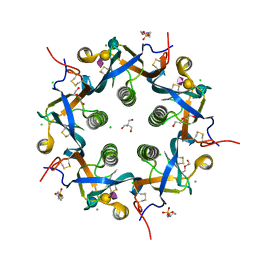 | | Crystal structure of 3'SL bound ArtB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ArtB protein, ... | | Authors: | Gao, X, Galan, J.E. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of host adaptation in the Salmonella typhoid toxin.
Nat Microbiol, 2, 2017
|
|
8FCE
 
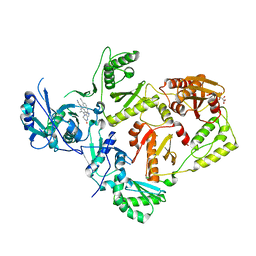 | | HIV-1 Reverse Transcriptase in complex with 7-membered bicyclic core NNRTI | | Descriptor: | 4-[(9-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
8FCD
 
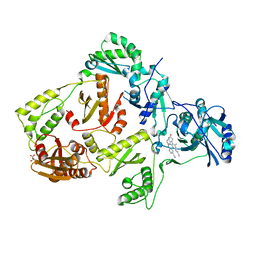 | | HIV-1 Reverse Transcriptase in complex with 6-membered bicyclic core NNRTI | | Descriptor: | 4-[(8-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-6-oxo-5,6,7,8-tetrahydropteridin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
8FCC
 
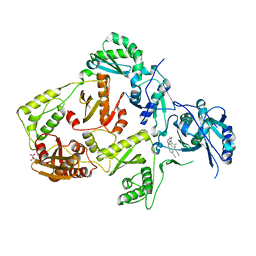 | | HIV-1 Reverse Transcriptase in complex with 5-membered bicyclic core NNRTI | | Descriptor: | 4-[(9-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-8-oxo-8,9-dihydro-7H-purin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
2DWU
 
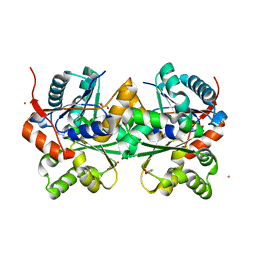 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | Authors: | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2006-08-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
2GZM
 
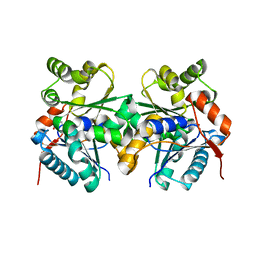 | | Crystal Structure of the Glutamate Racemase from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | May, M, Santarsiero, B.D, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design.
J.Mol.Biol., 371, 2007
|
|
4HND
 
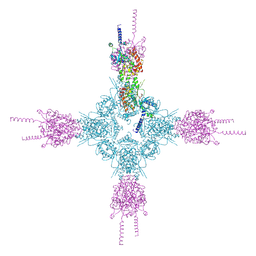 | | Crystal structure of the catalytic domain of Selenomethionine substituted human PI4KIIalpha in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
5WC2
 
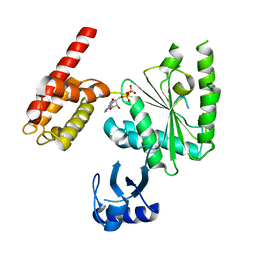 | | Crystal Structure of ADP-bound human TRIP13 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | Authors: | Jeong, B.-C, Luo, X. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Nat Commun, 8, 2017
|
|
4HNE
 
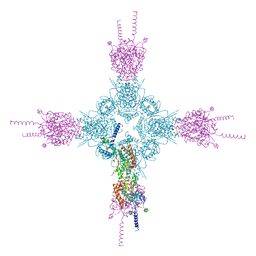 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
4K2C
 
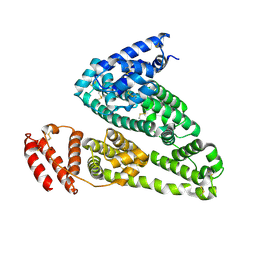 | | HSA Ligand Free | | Descriptor: | Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Huang, M. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin.
J. Biol. Chem., 288, 2013
|
|
4KGC
 
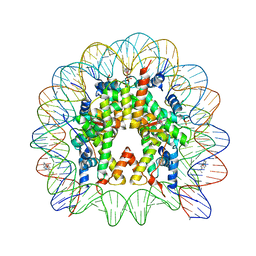 | | Nucleosome Core Particle Containing (ETA6-P-CYMENE)-(1, 2-ETHYLENEDIAMINE)-RUTHENIUM | | Descriptor: | (ethane-1,2-diamine-kappa~2~N,N')[(1,2,3,4,5,6-eta)-1-methyl-4-(propan-2-yl)cyclohexane-1,2,3,4,5,6-hexayl]ruthenium, DNA (145-mer), Histone H2A, ... | | Authors: | Adhireksan, Z, Davey, C.A. | | Deposit date: | 2013-04-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Ligand substitutions between ruthenium-cymene compounds can control protein versus DNA targeting and anticancer activity
Nat Commun, 5, 2014
|
|
8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
2I6B
 
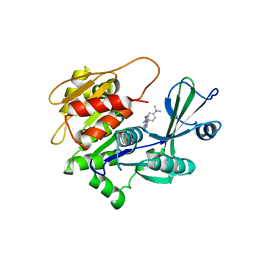 | | Human Adenosine Kinase in Complex with An Acetylinic Inhibitor | | Descriptor: | 5-[4-(DIMETHYLAMINO)PHENYL]-6-[(6-MORPHOLIN-4-YLPYRIDIN-3-YL)ETHYNYL]PYRIMIDIN-4-AMINE, Adenosine kinase | | Authors: | Muchmore, S.W. | | Deposit date: | 2006-08-28 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of human adenosine kinase inhibitor complexes reveal two distinct binding modes.
J.Med.Chem., 49, 2006
|
|
2I6A
 
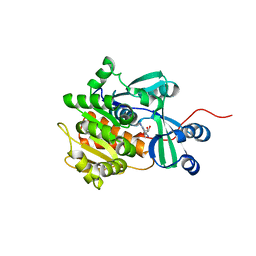 | | Human Adenosine Kinase in Complex With 5'-Deoxy-5-Iodotubercidin | | Descriptor: | 7-(5-DEOXY-BETA-D-RIBOFURANOSYL)-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, Adenosine kinase | | Authors: | Muchmore, S.W. | | Deposit date: | 2006-08-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human adenosine kinase inhibitor complexes reveal two distinct binding modes.
J.Med.Chem., 49, 2006
|
|
6ICT
 
 | | Structure of SETD3 bound to SAH and methylated actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
