8JYA
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYE
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYF
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with DMAPP | | Descriptor: | Butyrophylin 3, DIMETHYLALLYL DIPHOSPHATE, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYC
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with DMAPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8GVK
 
 | |
7FG9
 
 | | Alpha-1,2-glucosyltransferase_UDP_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7FGA
 
 | | Alpha-1,2-glucosyltransferase_UDP_sucrose_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-1)-alpha-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
5VJ8
 
 | |
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
8INP
 
 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT | | Descriptor: | Bc7OUGT, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
8ITA
 
 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT complexed with UDP and tectorigenin | | Descriptor: | 3-(4-hydroxyphenyl)-6-methoxy-5,7-bis(oxidanyl)chromen-4-one, Bc7OUGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
7Y77
 
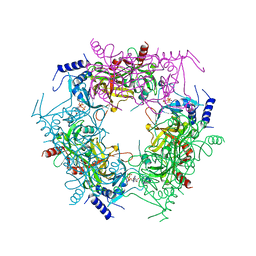 | | Crystal structure of rice NAL1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Yan, J.J, Guan, Z.Y, Yin, P, Xiong, L.Z. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Serine protease NAL1 exerts pleiotropic functions through degradation of TOPLESS-related corepressor in rice.
Nat.Plants, 9, 2023
|
|
5ZTM
 
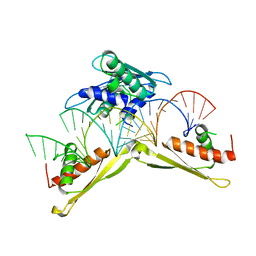 | | Crystal structure of MLE dsRBDs in complex with roX2 (R2H1) | | Descriptor: | Dosage compensation regulator, non-coding mRNA sequence roX2 | | Authors: | Lv, M.Q, Tang, Y.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights reveal the specific recognition of roX RNA by the dsRNA-binding domains of the RNA helicase MLE and its indispensable role in dosage compensation in Drosophila.
Nucleic Acids Res., 47, 2019
|
|
6KJY
 
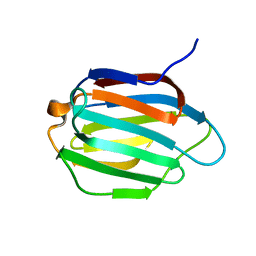 | | Galectin-13 variant C136S/C138S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
6KJW
 
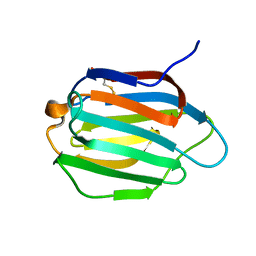 | | Galectin-13 variant C136S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
6KJX
 
 | | Galectin-13 variant C138S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
7EA8
 
 | |
7EA5
 
 | |
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7C5G
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5N
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.0 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5H
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 2.09 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5I
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 2.49 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5L
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
