7Y3C
 
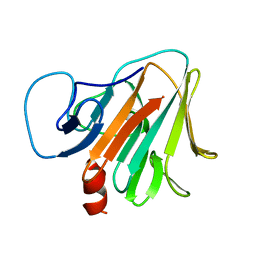 | | Crystal structure of TRIM7 bound to RACO-1 | | 分子名称: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | 著者 | Dong, C, Yan, X. | | 登録日 | 2022-06-10 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3B
 
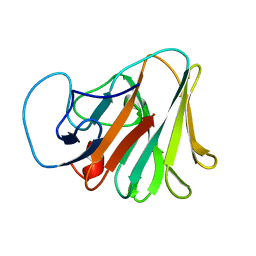 | | Crystal structure of TRIM7 bound to GN1 | | 分子名称: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | 著者 | Dong, C, Yan, X. | | 登録日 | 2022-06-10 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3A
 
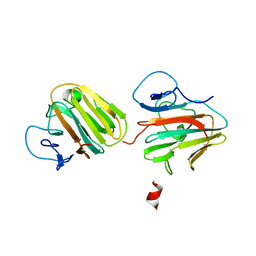 | | Crystal structure of TRIM7 bound to 2C | | 分子名称: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | 著者 | Dong, C, Yan, X. | | 登録日 | 2022-06-10 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U5V
 
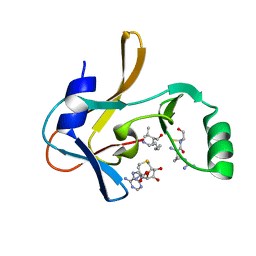 | | Crystal structure of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine and Borealin peptide | | 分子名称: | Borealin, Histone-lysine N-methyltransferase 2A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | An, S, Cho, U.S, Oh, H, Sha, L, Xu, J, Kim, S, Yang, W, An, W, Dou, Y. | | 登録日 | 2022-03-02 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Non-canonical MLL1 activity regulates centromeric phase separation and genome stability.
Nat.Cell Biol., 25, 2023
|
|
5WOE
 
 | |
5Y0G
 
 | |
5Y0X
 
 | |
5Y13
 
 | |
5Y0F
 
 | |
5Y12
 
 | |
5ZNP
 
 | |
5ZNR
 
 | |
5ZWZ
 
 | |
5ZWX
 
 | |
7DQA
 
 | | Cryo-EM structure of SARS-CoV2 RBD-ACE2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | 著者 | Wang, J, Lan, J, Wang, X.Q, Wang, H.W. | | 登録日 | 2020-12-22 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Reduced graphene oxide membrane as supporting film for high-resolution cryo-EM
Biophys Rep, 7, 2022
|
|
7YHP
 
 | |
7YHO
 
 | |
7YHQ
 
 | |
7YT9
 
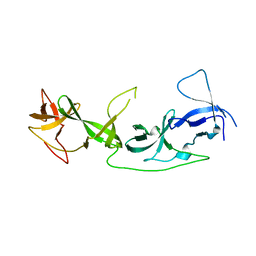 | | crystal structure of AGD1-4 of Arabidopsis AGDP3 | | 分子名称: | AGD1-4 of Arabidopsis AGDP3 | | 著者 | Zhou, X, Du, J. | | 登録日 | 2022-08-13 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-01-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
J Integr Plant Biol, 64, 2022
|
|
7YTA
 
 | |
7WHI
 
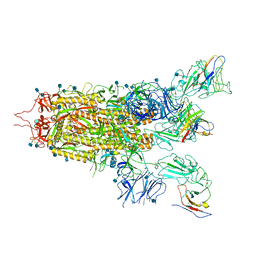 | | The state 2 complex structure of Omicron spike with Bn03 (2-up RBD, 4 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHK
 
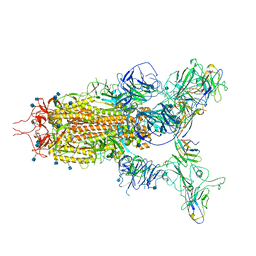 | | The state 3 complex structure of Omicron spike with Bn03 (2-up RBD, 5 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHJ
 
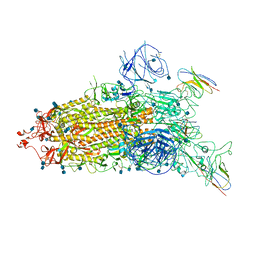 | | The state 1 complex structure of Omicron spike with Bn03 (1-up RBD, 3 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | 分子名称: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | 著者 | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | 登録日 | 2022-03-15 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
7U2X
 
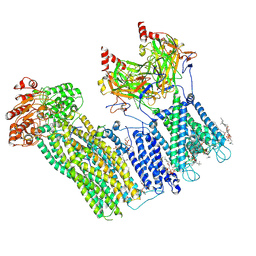 | | Cryo-EM structure of the pancreatic ATP-sensitive potassium channel in the presence of carbamazepine and ATP with Kir6.2-CTD in the down conformation | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Shyng, S.L, Sung, M.W, Driggers, C.M. | | 登録日 | 2022-02-25 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Ligand-mediated Structural Dynamics of a Mammalian Pancreatic K ATP Channel.
J.Mol.Biol., 434, 2022
|
|
