6OBW
 
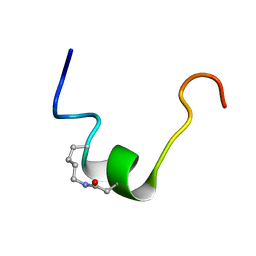 | | CSP1-cyc(K6D10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OC2
 
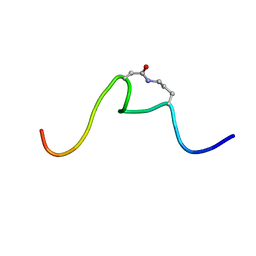 | | CSP1-cyc(Orn6D10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2JOB
 
 | | Solution structure of an antilipopolysaccharide factor from shrimp and its possible Lipid A binding site | | Descriptor: | antilipopolysaccharide factor | | Authors: | Yang, Y, Boze, H, Chemardin, P, Padilla, A, Moulin, G, Tassanakajon, A, Pugniere, M, Roquet, F, Gueguen, Y, Bachere, E, Aumelas, A. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: Model of the possible lipid A-binding site
Biopolymers, 91, 2009
|
|
6V1N
 
 | | CSP1-E1A-cyc(Dap6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-01-08 | | Last modified: | 2025-04-02 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2JZ1
 
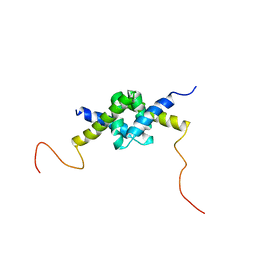 | | DSX_long | | Descriptor: | Protein doublesex | | Authors: | Yang, Y, Zhang, W, Bayrer, J.R, Weiss, M.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
J.Biol.Chem., 283, 2008
|
|
2KMM
 
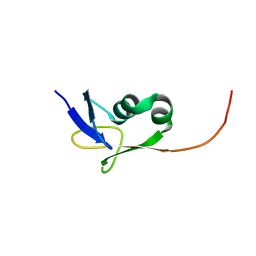 | | Solution NMR structure of the TGS domain of PG1808 from Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target PgR122A (418-481) | | Descriptor: | Guanosine-3',5'-bis(Diphosphate) 3'-pyrophosphohydrolase | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the TGS domain of PG1808 from
Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target
PgR122A (418-481)
To be Published
|
|
2KON
 
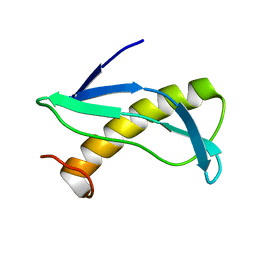 | | NMR solution structure of CV_2116 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvT4(1-82) | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Garcia, M, Yee, A, Arrowsmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of CV_2116 from Chromobacterium violaceum.Northeast Structural Genomics Consortium Target CvT4(1-82)
To be Published
|
|
6XLK
 
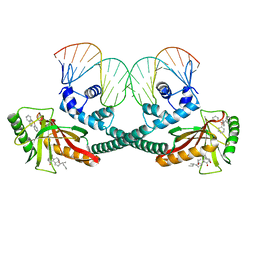 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-4nt | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLA
 
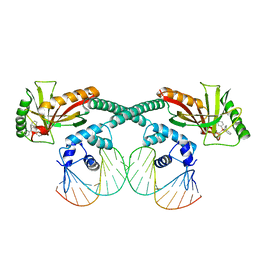 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | | Descriptor: | MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, synthetic non-template strand DNA (54-MER), ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL6
 
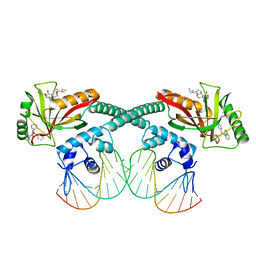 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPo | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLN
 
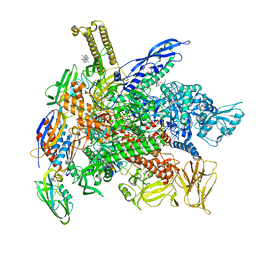 | | Cryo-EM structure of E. coli RNAP-DNA elongation complex 2 (RDe2) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLM
 
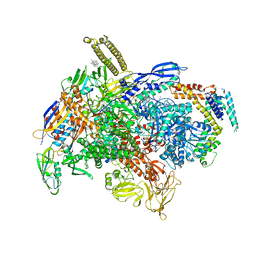 | | Cryo-EM structure of E.coli RNAP-DNA elongation complex 1 (RDe1) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLL
 
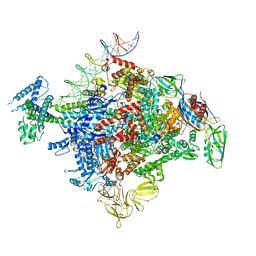 | | Cryo-EM structure of E. coli RNAP-promoter initial transcribing complex with 5-nt RNA transcript (RPitc-5nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL9
 
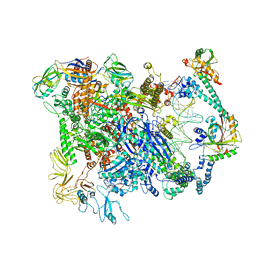 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 3-nt RNA transcript (EcmrR-RPitc-3nt) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLJ
 
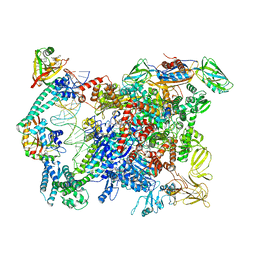 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 4-nt RNA transcript (EcmrR-RPitc-4nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL5
 
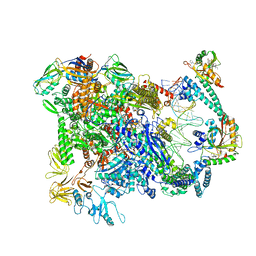 | | Cryo-EM structure of EcmrR-RNAP-promoter open complex (EcmrR-RPo) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
8QN7
 
 | | Amyloid-beta 40 type 1 filament from the leptomeninges of individual with Alzheimer's disease and cerebral amyloid angiopathy | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Yang, Y, Murzin, A.S, Peak-Chew, S.Y, Franco, C, Newell, K.L, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2023-09-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of A beta 40 filaments from the leptomeninges of individuals with Alzheimer's disease and cerebral amyloid angiopathy.
Acta Neuropathol Commun, 11, 2023
|
|
8QN6
 
 | | Amyloid-beta 40 type 2 filament from the leptomeninges of individual with Alzheimer's disease and cerebral amyloid angiopathy | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Yang, Y, Murzin, A.S, Peak-Chew, S.Y, Franco, C, Newell, K.L, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2023-09-25 | | Release date: | 2023-12-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structures of A beta 40 filaments from the leptomeninges of individuals with Alzheimer's disease and cerebral amyloid angiopathy.
Acta Neuropathol Commun, 11, 2023
|
|
7Q4M
 
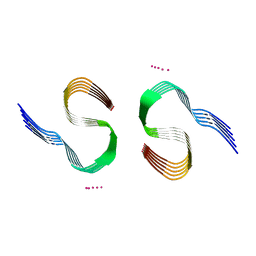 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
7Q4B
 
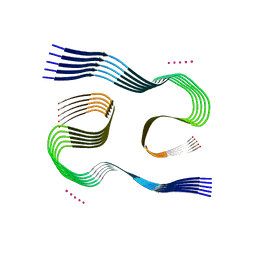 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
1L1C
 
 | | Structure of the LicT Bacterial Antiterminator Protein in Complex with its RNA Target | | Descriptor: | Transcription antiterminator licT, licT mRNA antiterminator hairpin | | Authors: | Yang, Y, Declerck, N, Manival, X, Aymerich, S, Kochoyan, M. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT.
EMBO J., 21, 2002
|
|
2RVJ
 
 | |
7K9C
 
 | |
7K9E
 
 | |
7K9D
 
 | |
