6K1M
 
 | | Engineered form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
6K1O
 
 | | Apo form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
6K1L
 
 | | E53A mutant of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
1P5T
 
 | | Crystal Structure of Dok1 PTB Domain | | Descriptor: | Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yuan, J, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.BIOL.CHEM., 279, 2004
|
|
1UEF
 
 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
3LZ6
 
 | | Guinea Pig 11beta hydroxysteroid dehydrogenase with PF-877423 | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-adamantan-2-yl-1-ethyl-D-prolinamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pauly, T.A. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The development and SAR of pyrrolidine carboxamide 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6LJ9
 
 | |
6LJB
 
 | |
6KZ6
 
 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
4R8Q
 
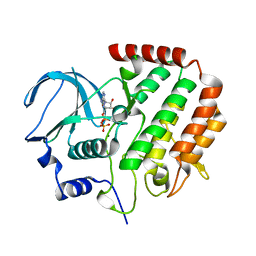 | |
7YHQ
 
 | |
7YHP
 
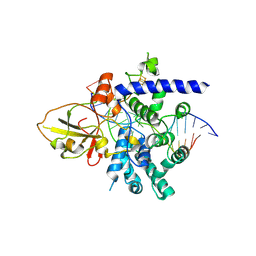 | |
7YHO
 
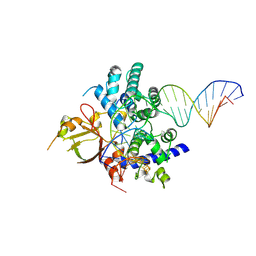 | |
6PAV
 
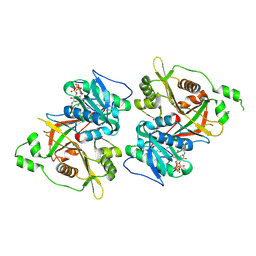 | |
6PAU
 
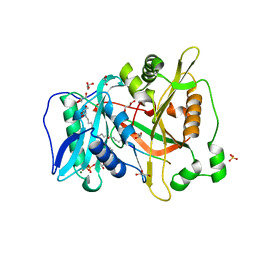 | |
2BX3
 
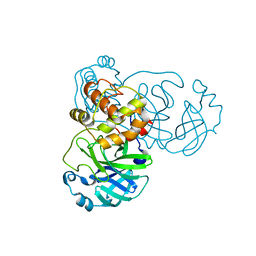 | |
2BX4
 
 | |
5HAD
 
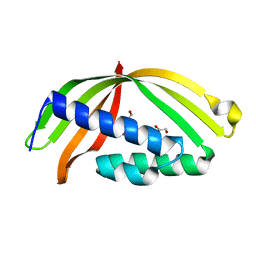 | |
5HZG
 
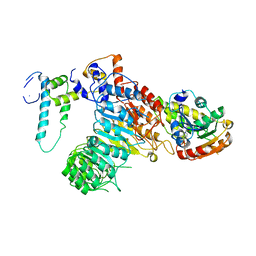 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
6A58
 
 | | Structure of histone demethylase REF6 | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
6A57
 
 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
5HYW
 
 | | The crystal structure of the D3-ASK1 complex | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
6A59
 
 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
1P5J
 
 | | Crystal Structure Analysis of Human Serine Dehydratase | | Descriptor: | L-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sun, L, Liu, Y, Rao, Z. | | Deposit date: | 2003-04-27 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of human serine dehydratase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
