2RUC
 
 | |
2RUD
 
 | |
6KVG
 
 | |
6Q7U
 
 | |
2DPF
 
 | | Crystal Structure of curculin1 homodimer | | 分子名称: | Curculin, SULFATE ION | | 著者 | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | 登録日 | 2006-05-11 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
8HT7
 
 | |
1SSK
 
 | | Structure of the N-terminal RNA-binding Domain of the SARS CoV Nucleocapsid Protein | | 分子名称: | Nucleocapsid protein | | 著者 | Huang, Q, Yu, L, Petros, A.M, Gunasekera, A, Liu, Z, Xu, N, Hajduk, P, Mack, J, Fesik, S.W, Olejniczak, E.T. | | 登録日 | 2004-03-24 | | 公開日 | 2004-06-08 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the N-Terminal RNA-Binding Domain of the SARS CoV Nucleocapsid Protein.
Biochemistry, 43, 2004
|
|
2A9H
 
 | | NMR structural studies of a potassium channel / charybdotoxin complex | | 分子名称: | Voltage-gated potassium channel, charybdotoxin | | 著者 | Yu, L, Sun, C, Song, D, Shen, J, Xu, N, Gunasekera, A, Hajduk, P.J, Olejniczak, E.T. | | 登録日 | 2005-07-11 | | 公開日 | 2006-01-10 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex.
Biochemistry, 44, 2005
|
|
4HMS
 
 | | Crystal structure of PhzG from Pseudomonas fluorescens 2-79 in complex with a second FMN in the substrate binding site | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | 著者 | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMV
 
 | | Crystal structure of PhzG from Pseudomonas fluorescens 2-79 in complex with tetrahydrophenazine-1-carboxylic acid after 5 days of soaking | | 分子名称: | (1R,10aS)-1,2,10,10a-tetrahydrophenazine-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, ... | | 著者 | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMW
 
 | |
4HMT
 
 | | Crystal structure of PhzG from Pseudomonas fluorescens 2-79 in complex with hexahydrophenazine-1,6-dicarboxylic acid | | 分子名称: | (1R,5aS,6R)-1,2,5,5a,6,7-hexahydrophenazine-1,6-dicarboxylic acid, FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, ... | | 著者 | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMU
 
 | | Crystal structure of PhzG from Pseudomonas fluorescens 2-79 in complex with tetrahydrophenazine-1-carboxylic acid after 1 day of soaking | | 分子名称: | (1R,10aS)-1,2,10,10a-tetrahydrophenazine-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, ... | | 著者 | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMX
 
 | | Crystal structure of PhzG from Burkholderia lata 383 in complex with tetrahydrophenazine-1-carboxylic acid | | 分子名称: | (1R,10aS)-1,2,10,10a-tetrahydrophenazine-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, Pyridoxamine 5'-phosphate oxidase | | 著者 | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1C9Q
 
 | |
1F9X
 
 | | AVERAGE NMR SOLUTION STRUCTURE OF THE BIR-3 DOMAIN OF XIAP | | 分子名称: | INHIBITOR OF APOPTOSIS PROTEIN XIAP, ZINC ION | | 著者 | Sun, C, Cai, M, Meadows, R.P, Fesik, S.W. | | 登録日 | 2000-07-11 | | 公開日 | 2001-07-11 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP.
J.Biol.Chem., 275, 2000
|
|
2RUQ
 
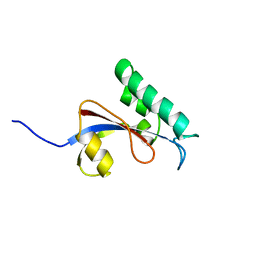 | |
2RUR
 
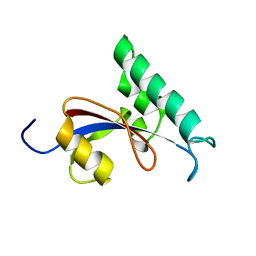 | |
6Q7W
 
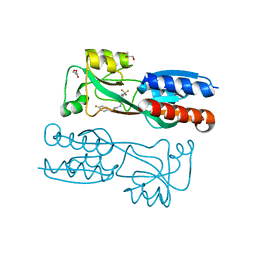 | |
6Q7V
 
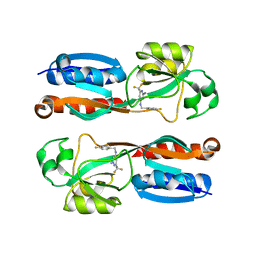 | |
5GS4
 
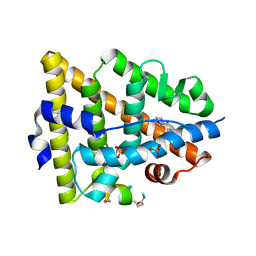 | | Crystal structure of estrogen receptor alpha in complex with a stabilized peptide antagonist | | 分子名称: | ARG-IAS-ILE-LEU-DNP-ARG-LEU-LEU-GLN, ESTRADIOL, Estrogen receptor, ... | | 著者 | Xie, M, Wang, T, Li, Z.-G. | | 登録日 | 2016-08-13 | | 公開日 | 2017-08-30 | | 最終更新日 | 2018-07-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of Inhibition of ER alpha-Coactivator Interaction by High-Affinity N-Terminus Isoaspartic Acid Tethered Helical Peptides
J. Med. Chem., 60, 2017
|
|
5GTR
 
 | |
6A37
 
 | |
5KD1
 
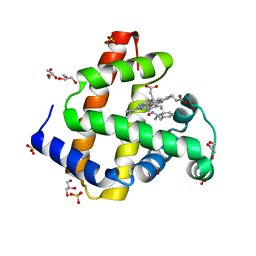 | | Sperm whale myoglobin H64A with nitrosoamphetamine | | 分子名称: | GLYCEROL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Wang, B, Guan, Y, Thomas, L.M, Richter-Addo, G.B. | | 登録日 | 2016-06-07 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Nitrosoamphetamine binding to myoglobin and hemoglobin: Crystal structure of the H64A myoglobin-nitrosoamphetamine adduct.
Nitric Oxide, 67, 2017
|
|
5YIM
 
 | |
