8J5U
 
 | | Crystal structure of Mycobacterium tuberculosis OppA complexed with an endogenous oligopeptide | | Descriptor: | Endogenous oligopeptide, Uncharacterized protein Rv1280c | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5R
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the resting state | | Descriptor: | IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, Putative peptide transport permease protein Rv1283c, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5S
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-catalytic intermediate state | | Descriptor: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5T
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the catalytic intermediate state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5Q
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-translocation state | | Descriptor: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3I7G
 
 | | MMP-13 in complex with a non zinc-chelating inhibitor | | Descriptor: | 5-(4-chlorophenyl)-N-[(1S)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]furan-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3I7I
 
 | | MMP-13 in complex with a non zinc-chelating inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N-[4-(5-{[(1S)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]carbamoyl}furan-2-yl)phenyl]-1-benzofuran-2-carboxamide, ... | | Authors: | Farrow, N.A, Margarit, S.M. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GI3
 
 | | Crystal structure of a N-Phenyl-N'-Naphthylurea analog in complex with p38 MAP kinase | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-tert-butyl-2-methoxy-3-[({4-[6-(morpholin-4-ylmethyl)pyridin-3-yl]naphthalen-1-yl}carbamoyl)amino]phenyl}methanesulfonamide | | Authors: | Qian, K.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and characterization of the N-phenyl-N'-naphthylurea class of p38 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5BOT
 
 | |
5XBL
 
 | | Structure of nuclease in complex with associated protein | | Descriptor: | Associated protein, CRISPR-associated endonuclease Cas9/Csn1, RNA (98-MER) | | Authors: | Dong, D, Guo, M, Wang, S, Zhu, Y, Huang, Z. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Nature, 546, 2017
|
|
5BOY
 
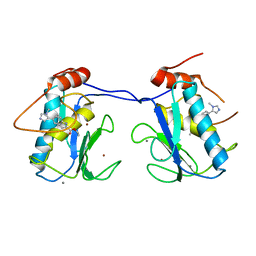 | |
5BPA
 
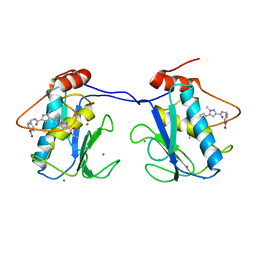 | |
7JU8
 
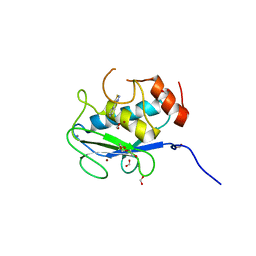 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
3QAH
 
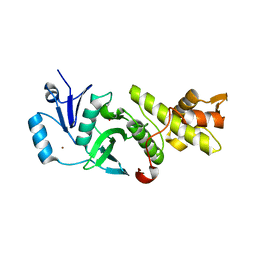 | | Crystal structure of apo-form human MOF catalytic domain | | Descriptor: | Probable histone acetyltransferase MYST1, ZINC ION | | Authors: | Sun, B, Tang, Q, Zhong, C, Ding, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274
Cell Res., 21, 2011
|
|
6IUP
 
 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6ITJ
 
 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6IUO
 
 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
8JA8
 
 | |
8JAC
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-16 | | Descriptor: | N-[[(2R,3S,4S,5R,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl]ethanamide, SULFATE ION, Trehalose-binding lipoprotein LpqY, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JA7
 
 | | Cryo-EM structure of Mycobacterium tuberculosis LpqY-SugABC in complex with trehalose | | Descriptor: | Trehalose import ATP-binding protein SugC, Trehalose transport system permease protein SugA, Trehalose transport system permease protein SugB, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAB
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-06 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-2-(fluoranylmethyl)-6-[(2~{R},3~{R},4~{S},5~{S},6~{S})-6-(fluoranylmethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-oxane-3,4,5-triol, SULFATE ION, Trehalose-binding lipoprotein LpqY | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAD
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-17 | | Descriptor: | BENZOIC ACID, SULFATE ION, Trehalose-binding lipoprotein LpqY, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JA9
 
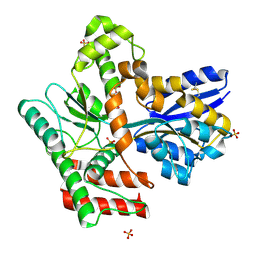 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-03 | | Descriptor: | SULFATE ION, Trehalose-binding lipoprotein LpqY, alpha-D-glucopyranose-(1-1)-(2~{S},3~{R},4~{S},5~{S},6~{S})-6-[(2-azanylhydrazinyl)methyl]oxane-2,3,4,5-tetrol | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAA
 
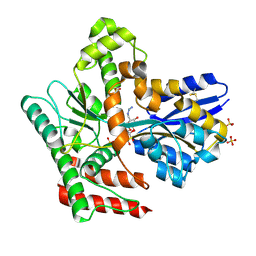 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-04 | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{S})-6-[(2-azanylhydrazinyl)methyl]oxane-2,3,4,5-tetrol, SULFATE ION, Trehalose-binding lipoprotein LpqY | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WIX
 
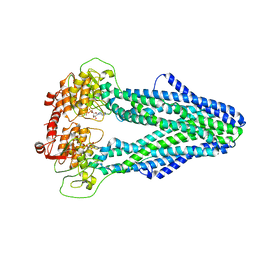 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
