3T6R
 
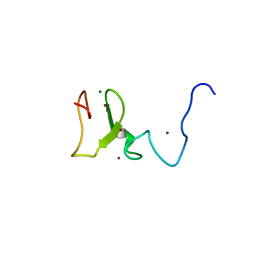 | | Structure of UHRF1 in complex with unmodified H3 N-terminal tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.1t N-terminal peptide, MAGNESIUM ION, ... | | Authors: | Xie, S, Jakoncic, J, Qian, C.M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail
J.Mol.Biol., 415, 2012
|
|
4XHU
 
 | |
4XHT
 
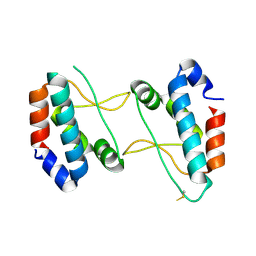 | |
4XHW
 
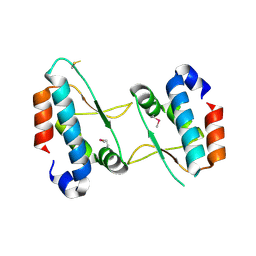 | |
4MQV
 
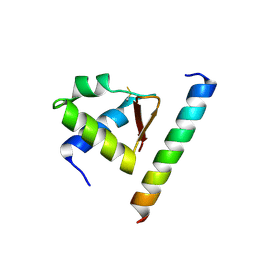 | | Crystal complex of Rpa32c and Smarcal1 N-terminus | | Descriptor: | Replication protein A 32 kDa subunit, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 | | Authors: | Xie, S, Qian, C.M. | | Deposit date: | 2013-09-16 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of RPA32 bound to the N-terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins
Febs J., 281, 2014
|
|
4ZDT
 
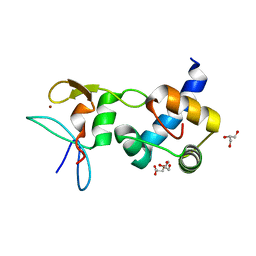 | | Crystal structure of the RING finger domain of Slx1 in complex with the C-terminal domain of Slx4 | | Descriptor: | GLYCEROL, SULFATE ION, Structure-specific endonuclease subunit slx1, ... | | Authors: | Lian, F.M, Xie, S, Qian, C.M. | | Deposit date: | 2015-04-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and SUMO binding of Slx1-Slx4 complex
Sci Rep, 6, 2016
|
|
9NPJ
 
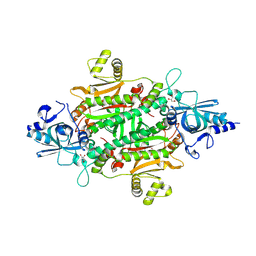 | | Structural studies of reaction hijacking inhibition of a malaria parasite aspartyl-tRNA synthetase. | | Descriptor: | 9-(5-O-{[(1R,2R)-2-amino-3-carboxy-1-hydroxypropyl]sulfamoyl}-beta-D-lyxofuranosyl)-2-chloro-9H-purin-6-amine, GLYCEROL, aspartate--tRNA ligase | | Authors: | Khandokar, Y, Ketprasit, K, Tai, C.W, Xie, S, Tilley, L, Griffin, M.D.W, Panjikar, S. | | Deposit date: | 2025-03-11 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural studies of reaction hijacking inhibition of a malaria parasite aspartyl-tRNA synthetase.
To Be Published
|
|
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
4GXO
 
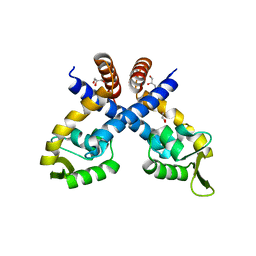 | | Crystal structure of Staphylococcus aureus protein SarZ mutant C13E | | Descriptor: | GLYCEROL, MarR family regulatory protein | | Authors: | Sun, F, Ding, Y, Ji, Q, Liang, Z, Deng, X, Wong, C.C, Yi, C, Zhang, L, Xie, S, Alvarez, S, Hicks, L.M, Luo, C, Jiang, H, Lan, L, He, C. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5TBM
 
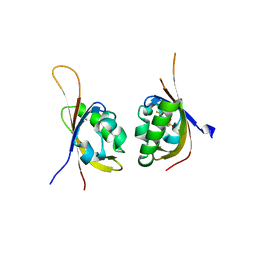 | | Crystal structure of PT2385 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Small-Molecule Antagonist of HIF2 alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma.
Cancer Res., 76, 2016
|
|
9LO7
 
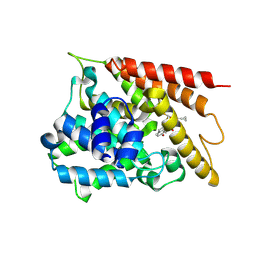 | | The crystal structure of PDE4D with 2317b | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huag, Y.-Y, Wu, D, Luo, H.-B. | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.20006251 Å) | | Cite: | Structure-Based Optimization of Moracin M as Potent and Selective PDE4 Inhibitors with Antipsoriasis Effects.
J.Med.Chem., 68, 2025
|
|
8T1R
 
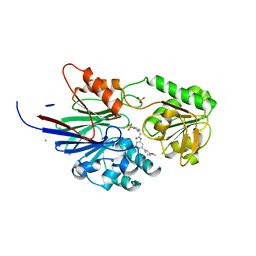 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
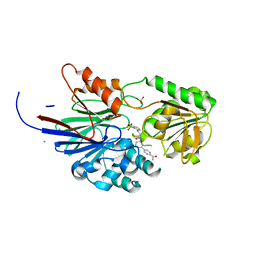 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
5WYM
 
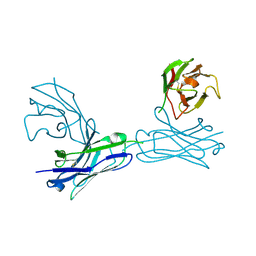 | | Crystal structure of an anti-connexin26 scFv | | Descriptor: | anti-connexin26 scFv,Ig heavy chain,Linker,anti-connexin26 scFv,Ig light chain | | Authors: | Li, S, Xu, L. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Characterization of a Human Monoclonal Antibody that Modulates Mutant Connexin 26 Hemichannels Implicated in Deafness and Skin Disorders
Front Mol Neurosci, 10, 2017
|
|
8ZQU
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 2-6 | | Descriptor: | (3~{Z})-3-[[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylidene]-6-oxidanyl-1-benzofuran-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-06-03 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.00004554 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
8ZQW
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 2-9 | | Descriptor: | (3~{Z})-3-[[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylidene]-6-oxidanyl-1~{H}-indol-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-06-03 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.100019 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
8ZQ2
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 2-1 | | Descriptor: | (3~{Z})-3-[(3-ethoxy-4-methoxy-phenyl)methylidene]-6-oxidanyl-1-benzofuran-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-05-31 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.10001445 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
8ZQ1
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 1-12 | | Descriptor: | (3~{E})-3-[(3,4-dimethoxyphenyl)methylidene]-6-oxidanyl-1-benzofuran-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-05-31 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.200001 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
8JKZ
 
 | |
8JL0
 
 | | Cryo-EM structure of the prokaryotic SPARSA system complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain protein, ... | | Authors: | Xu, X, Zhen, X, Long, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antiphage immunity generated by a prokaryotic Argonaute-associated SPARSA system.
Nat Commun, 15, 2024
|
|
5Z65
 
 | | Crystal structure of porcine aminopeptidase N ectodomain in functional form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, Y.G, Li, R, Jiang, L.G, Qiao, S.L, Zhang, G.P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Characterization of the interaction between recombinant porcine aminopeptidase N and spike glycoprotein of porcine epidemic diarrhea virus.
Int. J. Biol. Macromol., 117, 2018
|
|
8K9D
 
 | | Structure of human Caprin-2 HR1 domain | | Descriptor: | Caprin-2 | | Authors: | Song, X.M. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the Caprin-2 HR1 domain in canonical Wnt signaling.
J.Biol.Chem., 300, 2024
|
|
2M6X
 
 | |
5GJK
 
 | | Crystal Structure of BAF47 and BAF155 Complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, SWI/SNF complex subunit SMARCC1, ... | | Authors: | Yan, L, Qian, C. | | Deposit date: | 2016-06-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural Insights into BAF47 and BAF155 Complex Formation.
J. Mol. Biol., 429, 2017
|
|
5YDR
 
 | | Structure of DNMT1 RFTS domain in complex with ubiquitin | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, PHOSPHATE ION, Polyubiquitin-B, ... | | Authors: | Qian, C. | | Deposit date: | 2017-09-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.
Nucleic Acids Res., 46, 2018
|
|
