8RGI
 
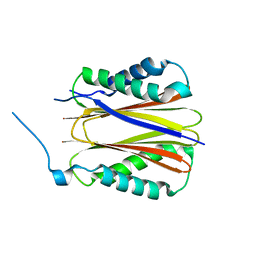 | | Structure of DYNLT1:DYNLT2B (TCTEX1:TCTEX1D2) heterodimer. | | Descriptor: | Dynein light chain Tctex-type 1, Dynein light chain Tctex-type protein 2B | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGG
 
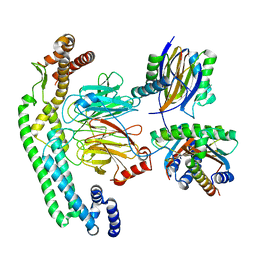 | | Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Dynein light chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Mladenov, M, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGH
 
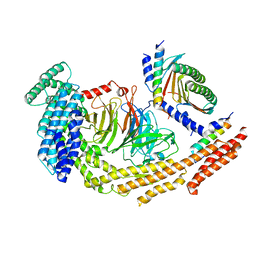 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
6RMH
 
 | | The Rigid-body refined model of the normal Huntingtin. | | Descriptor: | Huntingtin | | Authors: | Jung, T, Tamo, G, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2019-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
4BLB
 
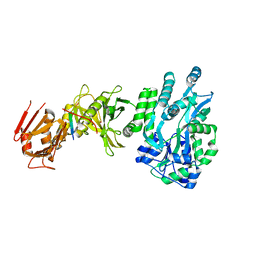 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6YEJ
 
 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | Descriptor: | Huntingtin | | Authors: | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2020-03-24 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (18.200001 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
4BLD
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI3p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, TRANSCRIPTIONAL ACTIVATOR GLI3, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLA
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BL9
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BL8
 
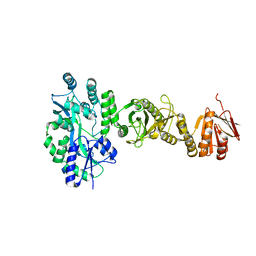 | | Crystal structure of full-length human Suppressor of fused (SUFU) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Karlstrom, M, Finta, C, Cherry, A.L, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3R6G
 
 | |
3R7S
 
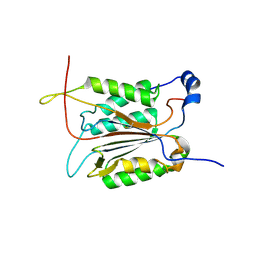 | | Crystal Structure of Apo Caspase2 | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18 | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R7N
 
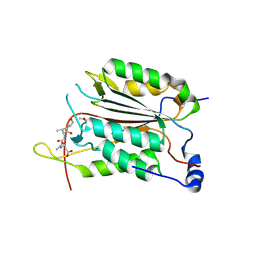 | | Caspase-2 bound with two copies of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R5J
 
 | |
3R6L
 
 | | Caspase-2 T380A bound with Ac-VDVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)VDVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R7B
 
 | | Caspase-2 bound to one copy of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
6G3X
 
 | |
6G3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | Descriptor: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
