1SL4
 
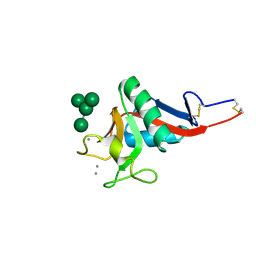 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1D2N
 
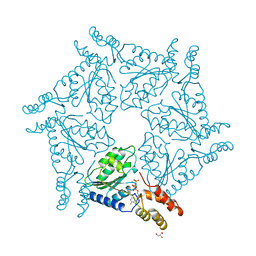 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
2FO0
 
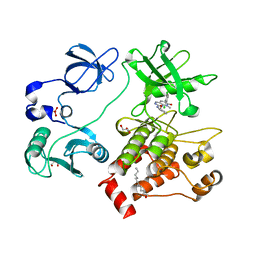 | | Organization of the SH3-SH2 Unit in Active and Inactive Forms of the c-Abl Tyrosine Kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Seeliger, M, Davies, J.M, Weis, W.I, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase.
Mol.Cell, 21, 2006
|
|
1DOV
 
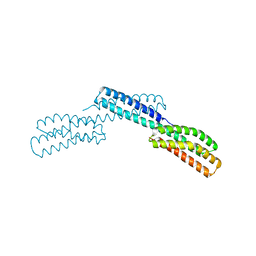 | |
1DK8
 
 | | CRYSTAL STRUCTURE OF THE RGS-HOMOLOGOUS DOMAIN OF AXIN | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, AXIN, GLYCEROL, ... | | Authors: | Spink, K.E, Polakis, P, Weis, W.I. | | Deposit date: | 1999-12-06 | | Release date: | 2000-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis of the Axin-adenomatous polyposis coli interaction.
EMBO J., 19, 2000
|
|
1DOW
 
 | |
1EGI
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
1EMU
 
 | |
1EGG
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
1FIH
 
 | | N-ACETYLGALACTOSAMINE BINDING MUTANT OF MANNOSE-BINDING PROTEIN A (QPDWG-HDRPY), COMPLEX WITH N-ACETYLGALACTOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Feinberg, H, Torgerson, D, Drickamer, K, Weis, W.I. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of pH-dependent N-acetylgalactosamine binding by a functional mimic of the hepatocyte asialoglycoprotein receptor.
J.Biol.Chem., 275, 2000
|
|
1FIF
 
 | | N-ACETYLGALACTOSAMINE-SELECTIVE MUTANT OF MANNOSE-BINDING PROTEIN-A (QPDWG-HDRPY) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Feinberg, H, Torgersen, D, Drickamer, K, Weis, W.I. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of pH-dependent N-acetylgalactosamine binding by a functional mimic of the hepatocyte asialoglycoprotein receptor.
J.Biol.Chem., 275, 2000
|
|
5XA5
 
 | | Crystal structure of HMP-1-HMP-2 complex | | Descriptor: | Alpha-catenin-like protein hmp-1, Beta-catenin-like protein hmp-2 | | Authors: | Shao, X, Kang, H, Weis, W.I, Hardin, J, Choi, H.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cell-cell adhesion in metazoans relies on evolutionarily conserved features of the alpha-catenin· beta-catenin-binding interface.
J.Biol.Chem., 292, 2017
|
|
5X7D
 
 | | Structure of beta2 adrenoceptor bound to carazolol and an intracellular allosteric antagonist | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liu, X, Ahn, S, Kahsai, A.W, Meng, K.-C, Latorraca, N.R, Pani, B, Venkatakrishnan, A.J, Masoudi, A, Weis, W.I, Dror, R.O, Chen, X, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2017-02-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Mechanism of intracellular allosteric beta 2AR antagonist revealed by X-ray crystal structure.
Nature, 548, 2017
|
|
3P5F
 
 | | Structure of the carbohydrate-recognition domain of human Langerin with man2 (Man alpha1-2 Man) | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-D-mannopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7501 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5H
 
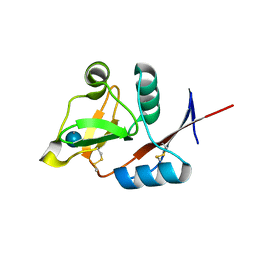 | | Structure of the carbohydrate-recognition domain of human Langerin with Laminaritriose | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, beta-D-glucopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6051 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P0G
 
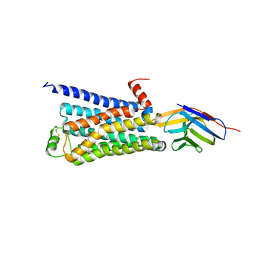 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
3P5E
 
 | | Structure of the carbohydrate-recognition domain of human Langerin with man4 (Man alpha1-3(Man alpha1-6)Man alpha1-6Man) | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-D-mannopyranose | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7012 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5G
 
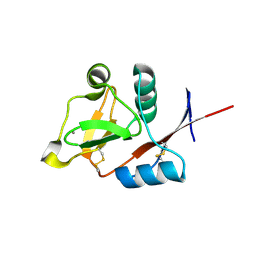 | | Structure of the carbohydrate-recognition domain of human Langerin with Blood group B trisaccharide (Gal alpha1-3(Fuc alpha1-2)Gal) | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-L-fucopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6027 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5I
 
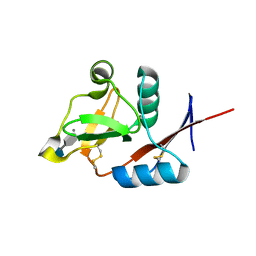 | | Structure of the carbohydrate-recognition domain of human Langerin with 6-SO4-Gal-GlcNAc | | Descriptor: | 6-O-sulfo-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5D
 
 | | Structure of the carbohydrate-recognition domain of human Langerin with man5 (Man alpha1-3(Man alpha1-6)Man alpha1-6)(Man- alpha1-3)Man | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-D-mannopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8013 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
4YUH
 
 | | Multiconformer synchrotron model of CypA at 150 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YUO
 
 | | High-resolution multiconformer synchrotron model of CypA at 273 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YUJ
 
 | | Multiconformer synchrotron model of CypA at 240 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YUN
 
 | | Multiconformer synchrotron model of CypA at 310 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
