5Y4D
 
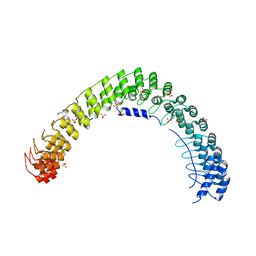 | | Crystal Structure of AnkB Ankyrin Repeats in Complex with AnkR/AnkB Chimeric Autoinhibition Segment | | Descriptor: | Ankyrin-1,Ankyrin-2,Ankyrin-2, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4F
 
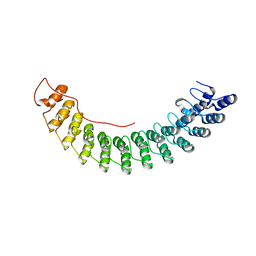 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | Descriptor: | ACETATE ION, Ankyrin-2, CALCIUM ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
6C1R
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist avacopan | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MALONATE ION, OLEIC ACID, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C1Q
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, PMX53, Soluble cytochrome b562, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7YV9
 
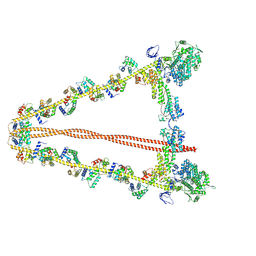 | |
7Y8W
 
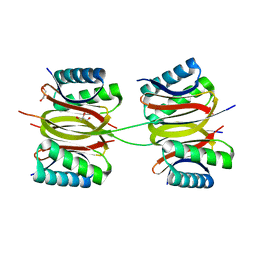 | | Crystal structure of DLC-1/SAO-1 complex | | Descriptor: | Dynein light chain 1, cytoplasmic, GLYCEROL, ... | | Authors: | Yan, H, Zhao, C, Wei, Z, Yu, C. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.
Cell Death Discov, 8, 2022
|
|
6IUI
 
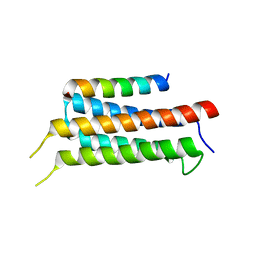 | | Crystal structure of GIT1 PBD domain in complex with Paxillin LD4 motif | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
6IUH
 
 | | Crystal structure of GIT1 PBD domain in complex with Liprin-alpha2 | | Descriptor: | ARF GTPase-activating protein GIT1, IODIDE ION, Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
6IXQ
 
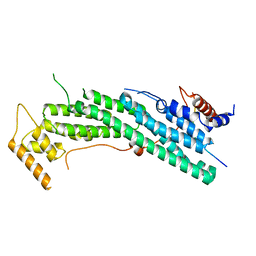 | | Structure of Myo2-GTD in complex with Smy1 | | Descriptor: | Kinesin-related protein SMY1, Myosin-2 | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6IXR
 
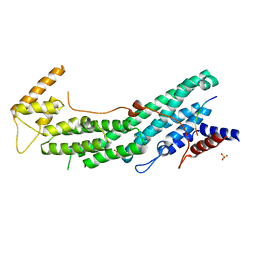 | | Structure of Myo2-GTD in complex with Inp2 | | Descriptor: | Inheritance of peroxisomes protein 2, Myosin-2, SULFATE ION | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6IXO
 
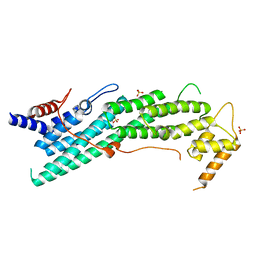 | | Apo structure of Myo2-GTD | | Descriptor: | CHLORIDE ION, Myosin-2, SULFATE ION | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6IXP
 
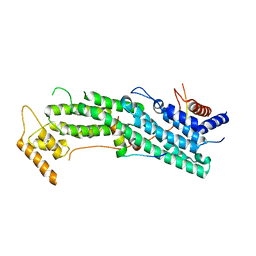 | | Structure of Myo2-GTD in complex with Mmr1 | | Descriptor: | Mitochondrial MYO2 receptor-related protein 1, Myosin-2 | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.733 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6KR4
 
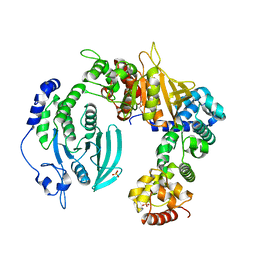 | | Crystal structure of the liprin-alpha3_SAM123/LAR_D1D2 complex | | Descriptor: | HEXAETHYLENE GLYCOL, Liprin-alpha-3, PHOSPHATE ION, ... | | Authors: | Xie, X, Liang, M, Luo, L, Wei, Z. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of liprin-alpha-promoted LAR-RPTP clustering for modulation of phosphatase activity.
Nat Commun, 11, 2020
|
|
6QSI
 
 | |
7ORX
 
 | |
3F1S
 
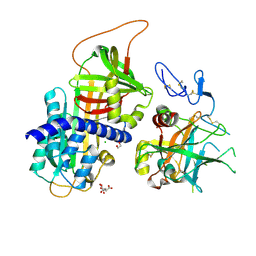 | | Crystal structure of Protein Z complexed with protein Z-dependent inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, A. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of protein Z-dependent inhibitor complex shows how protein Z functions as a cofactor in the membrane inhibition of factor X.
Blood, 2009
|
|
2KLQ
 
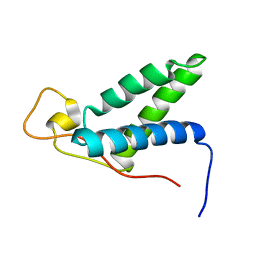 | | The solution structure of CBD of human MCM6 | | Descriptor: | DNA replication licensing factor MCM6 | | Authors: | Liu, C. | | Deposit date: | 2009-07-08 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6
J.Biol.Chem., 285, 2010
|
|
2A9J
 
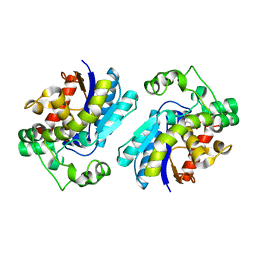 | |
5INW
 
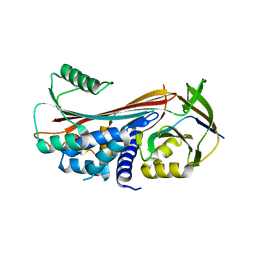 | | Structure of reaction loop cleaved lamprey angiotensinogen | | Descriptor: | C-terminal peptide of Putative angiotensinogen, Putative angiotensinogen, SULFATE ION | | Authors: | Wei, H, Zhou, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heparin Binds Lamprey Angiotensinogen and Promotes Thrombin Inhibition through a Template Mechanism
J.Biol.Chem., 291, 2016
|
|
7MWY
 
 | |
7MWZ
 
 | |
2ZMV
 
 | | Crystal structure of Synbindin | | Descriptor: | Trafficking protein particle complex subunit 4 | | Authors: | Fan, F. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human synbindin reveals two conformations of longin domain
Biochem.Biophys.Res.Commun., 378, 2009
|
|
4ZRJ
 
 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4ZRI
 
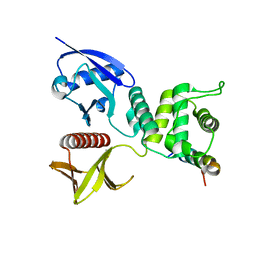 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
2LW9
 
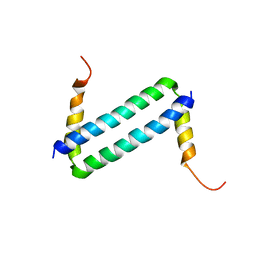 | | NMR solution structure of Myo10 anti-CC | | Descriptor: | Unconventionnal myosin-X | | Authors: | Ye, F, Lu, Q, Zhang, M. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-26 | | Last modified: | 2013-06-12 | | Method: | SOLUTION NMR | | Cite: | Antiparallel coiled-coil-mediated dimerization of myosin X
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
