6ON1
 
 | | A resting state structure of L-DOPA dioxygenase from Streptomyces sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, L-DOPA dioxygenase | | Authors: | Wang, Y, Shin, I, Fu, Y, Colabroy, K, Liu, A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal Structures of L-DOPA Dioxygenase fromStreptomyces sclerotialus.
Biochemistry, 58, 2019
|
|
3KCU
 
 | | Structure of formate channel | | Descriptor: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
3KCV
 
 | | Structure of formate channel | | Descriptor: | Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
9IOX
 
 | | Cryo-EM structure of a TEF30-associated intermediate PSII core dimer complex, type II, from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, Y, Wang, C, Li, A, Liu, Z. | | Deposit date: | 2024-07-09 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TEF30 fulfills versatile functions in the photosystem II repair process
To Be Published
|
|
9IMN
 
 | | Cryo-EM structure of a TEF30-associated intermediate PSII core dimer complex, type I, from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, Y, Wang, C, Li, A, Liu, Z. | | Deposit date: | 2024-07-03 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | TEF30 fulfills versatile functions in the photosystem II repair process
To Be Published
|
|
9IIU
 
 | | Cryo-EM structure of an TEF30-associated intermediate PSII core complex from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, Y, Wang, C, Li, A, Liu, Z. | | Deposit date: | 2024-06-21 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | TEF30 fulfills versatile functions in the photosystem II repair process
To Be Published
|
|
9IS4
 
 | | Cryo-EM structure of a TEF30-associated intermediate C2S-type PSII-LHCII supercomplex from Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wang, Y, Wang, C, Li, A, Liu, Z. | | Deposit date: | 2024-07-16 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | TEF30 fulfills versatile functions in the photosystem II repair process
To Be Published
|
|
7REI
 
 | | The crystal structure of nickel bound human ADO C18S C239S variant | | Descriptor: | 2-aminoethanethiol dioxygenase, GLYCEROL, NICKEL (II) ION | | Authors: | Wang, Y, Shin, I, Li, J, Liu, A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human cysteamine dioxygenase provides a structural rationale for its function as an oxygen sensor.
J.Biol.Chem., 297, 2021
|
|
6VI8
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a superoxo bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VI6
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a substrate monodentately bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VI7
 
 | | Probing extradiol dioxygenase mechanism in NAD(+) biosynthesis by viewing reaction cycle intermediates - a substrate bidentately bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.617 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9K3Z
 
 | |
9K40
 
 | |
9K41
 
 | |
9K42
 
 | |
9K43
 
 | |
8GAP
 
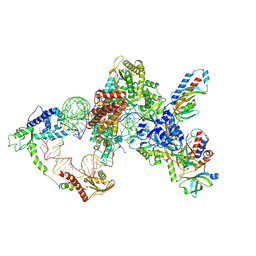 | | Structure of LARP7 protein p65-telomerase RNA complex in telomerase | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | Wang, Y, He, Y, Wang, Y, Yang, Y, Singh, M, Eichhorn, C.D, Zhou, Z.H, Feigon, J. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LARP7 Protein p65-telomerase RNA Complex in Telomerase Revealed by Cryo-EM and NMR.
J.Mol.Biol., 435, 2023
|
|
9IVD
 
 | |
3TDL
 
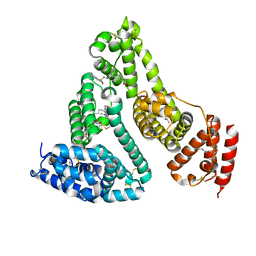 | | Structure of human serum albumin in complex with DAUDA | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | Deposit date: | 2011-08-11 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
9IID
 
 | | Crystal structure of PgAfp | | Descriptor: | Antifungal protein B | | Authors: | Wang, Y. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The role of Npt1 in regulating antifungal protein activity in filamentous fungi.
Nat Commun, 16, 2025
|
|
3K62
 
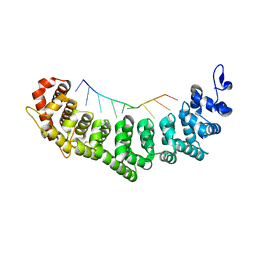 | | Crystal structure of FBF-2/gld-1 FBEb complex | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*UP*AP*UP*C)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K5Q
 
 | | Crystal structure of FBF-2/FBE complex | | Descriptor: | 5'-R(P*UP*GP*UP*AP*CP*UP*AP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K5Z
 
 | | Crystal structure of FBF-2/gld-1 FBEa G4A mutant complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*CP*CP*AP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K64
 
 | | Crystal structure of FBF-2/fem-3 PME complex | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*CP*AP*UP*U)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5GSW
 
 | | Crystal structure of EV71 3C in complex with N69S 1.8k | | Descriptor: | 3C protein, ~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]propan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wang, Y. | | Deposit date: | 2016-08-17 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the Enterovirus 71 3C Protease in Complex with NK-1.8k and Indications for the Development of Antienterovirus Protease Inhibitor
Antimicrob. Agents Chemother., 61, 2017
|
|
