4P69
 
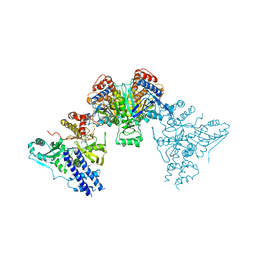 | | Acek (D477A) ICDH complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NADP], ... | | 著者 | Jimin, Z, Nan, W, Shu, W, Zongchao, J. | | 登録日 | 2014-03-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The phosphatase mechanism of bifunctional kinase/phosphatase AceK.
Chem. Commun. (Camb.), 50, 2014
|
|
7DRG
 
 | | Pseudomonas aeruginosa T6SS protein PA0821 | | 分子名称: | PA0821 | | 著者 | She, Z. | | 登録日 | 2020-12-28 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Pseudomonas aeruginosa PAAR2 cluster encodes a putative VRR-NUC domain-containing effector.
Febs J., 288, 2021
|
|
4WVD
 
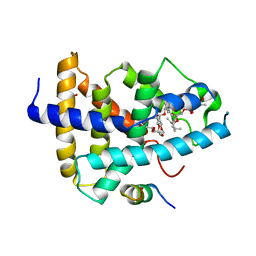 | | Identification of a novel FXR ligand that regulates metabolism | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Bile acid receptor, FORMIC ACID, ... | | 著者 | Wang, R, Li, Y. | | 登録日 | 2014-11-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism.
Nat Commun, 4, 2013
|
|
4W2R
 
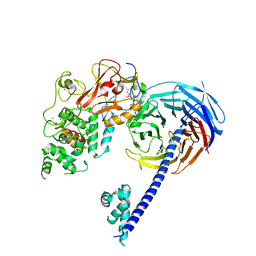 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | 分子名称: | 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | 著者 | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | 登録日 | 2017-09-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
7M4T
 
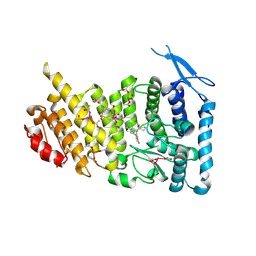 | | Menin bound to M-1121 | | 分子名称: | Menin, methyl {(1S,2R)-2-[(1S)-2-(azetidin-1-yl)-1-(3-fluorophenyl)-1-{1-[(3-methoxy-1-{4-[(1S,4S)-5-propanoyl-2,5-diazabicyclo[2.2.1]heptane-2-sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}ethyl]cyclopentyl}carbamate, praseodymium triacetate | | 著者 | Stuckey, J. | | 登録日 | 2021-03-22 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Discovery of M-1121 as an Orally Active Covalent Inhibitor of Menin-MLL Interaction Capable of Achieving Complete and Long-Lasting Tumor Regression.
J.Med.Chem., 64, 2021
|
|
8BNS
 
 | |
8FH4
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-13 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.827 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | 分子名称: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP3
 
 | | PKCeta kinase domain in complex with compound 11 | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FJZ
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-20 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
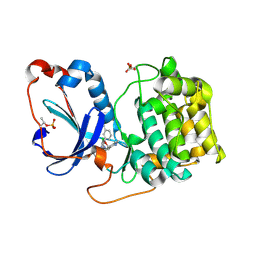 | | PKCeta kinase domain in complex with compound 2 | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
6OT1
 
 | |
6OSY
 
 | |
6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
8T5C
 
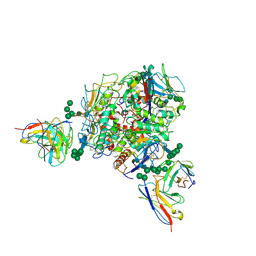 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-06-13 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
6C7R
 
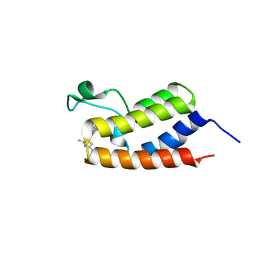 | | BRD4 BD1 in complex with compound CF53 | | 分子名称: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2018-01-23 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | 分子名称: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2018-05-31 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
4XGY
 
 | | GFP based antibody (fluorobody) | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Green fluorescent protein, mAb LCDR3 | | 著者 | Shi, N, Chen, Y.G, Wang, S.H. | | 登録日 | 2015-01-03 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.494 Å) | | 主引用文献 | The structure of a GFP-based antibody (fluorobody) to TLH, a toxin from Vibrio parahaemolyticus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8ERC
 
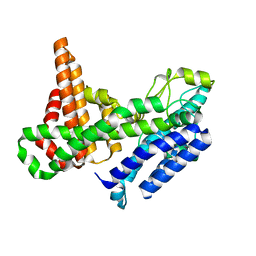 | |
1T1K
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ALA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | 分子名称: | Insulin | | 著者 | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | 登録日 | 2004-04-16 | | 公開日 | 2004-08-10 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
1T1Q
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ABA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | 分子名称: | Insulin, insulin | | 著者 | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | 登録日 | 2004-04-16 | | 公開日 | 2004-08-10 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
