6KH5
 
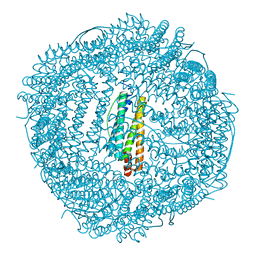 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | 分子名称: | FE (III) ION, Ferritin, NICKEL (II) ION | | 著者 | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | 登録日 | 2019-07-12 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.294 Å) | | 主引用文献 | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
5A8H
 
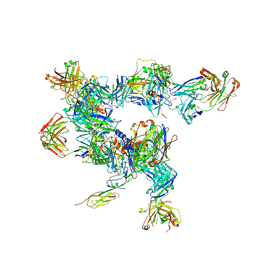 | | cryo-ET subtomogram averaging of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195 VARIANT G52K5, ... | | 著者 | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | 登録日 | 2015-07-15 | | 公開日 | 2015-08-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env.
Cell, 162, 2015
|
|
5A7X
 
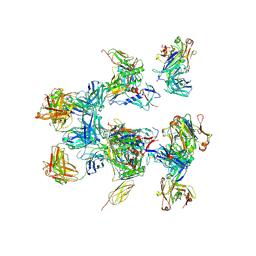 | | negative stain EM of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195, ... | | 著者 | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | 登録日 | 2015-07-10 | | 公開日 | 2015-08-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (17 Å) | | 主引用文献 | Broadly Neutralizing Antibody 8Anc195 Recognizes Closed and Open States of HIV-1 Env.
Cell(Cambridge,Mass.), 162, 2015
|
|
8T8M
 
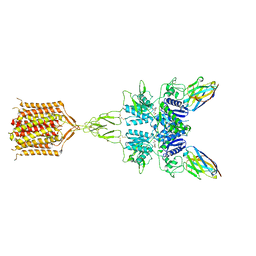 | | Quis-bound intermediate mGlu5 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nb43 | | 著者 | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | 登録日 | 2023-06-22 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Step-wise activation of a Family C GPCR.
Biorxiv, 2023
|
|
8T6J
 
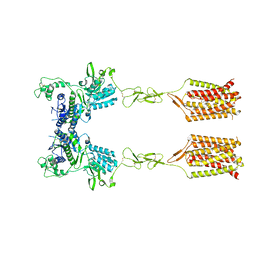 | |
8T7H
 
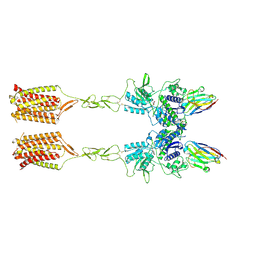 | | Quis-bound intermediate mGlu5 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | 著者 | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | 登録日 | 2023-06-20 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Step-wise activation of a Family C GPCR.
Biorxiv, 2023
|
|
8TAO
 
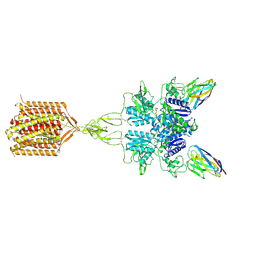 | | Quis and CDPPB bound active mGlu5 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5, ... | | 著者 | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | 登録日 | 2023-06-27 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Step-wise activation of a Family C GPCR.
Biorxiv, 2023
|
|
4LH6
 
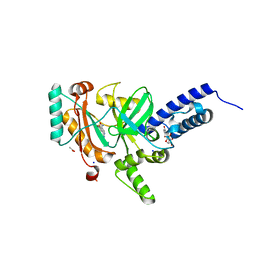 | | Crystal structure of a LigA inhibitor | | 分子名称: | 4-amino-2-bromothieno[3,2-c]pyridine-7-carboxamide, ACETATE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ... | | 著者 | Benenato, K, Wang, H, Mcguire, H.M, Davis, H, Gao, N, Prince, D.B, Jahic, H, Stokes, S.S, Boriack-Sjodin, P.A. | | 登録日 | 2013-06-30 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3TDL
 
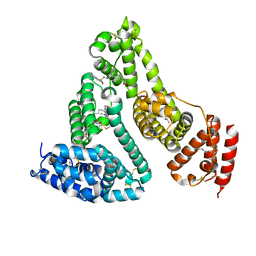 | | Structure of human serum albumin in complex with DAUDA | | 分子名称: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | 著者 | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | 登録日 | 2011-08-11 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
1Q2I
 
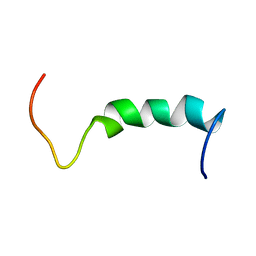 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | 分子名称: | PNC27 | | 著者 | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | 登録日 | 2003-07-24 | | 公開日 | 2004-03-16 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1Q2F
 
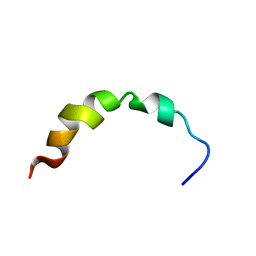 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | 分子名称: | PNC27 | | 著者 | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | 登録日 | 2003-07-24 | | 公開日 | 2004-03-16 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
8HFQ
 
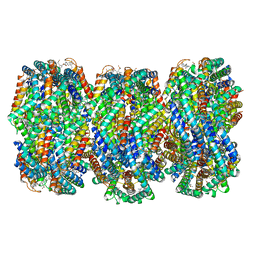 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | 分子名称: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | 著者 | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | 登録日 | 2022-11-11 | | 公開日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
2QGM
 
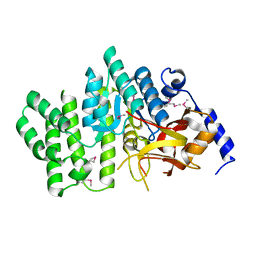 | | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136. | | 分子名称: | Succinoglycan biosynthesis protein | | 著者 | Kuzin, A.P, Abashidze, M, Jayaraman, S, Wang, H, Fang, Y, Maglaqui, M, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-29 | | 公開日 | 2007-07-24 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136.
To be Published
|
|
3QFI
 
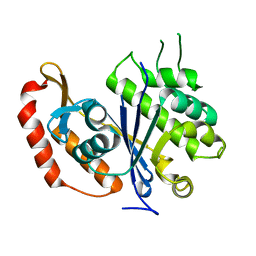 | | X-ray crystal structure of transcriptional regulator (EF0465) from Enterococcus faecalis, Northeast Structural Genomics Consortium Target EfR190 | | 分子名称: | Transcriptional regulator | | 著者 | Forouhar, F, Neely, H, Seetharaman, J, Wang, H, Ciccosanti, C, Mao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hun, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-01-21 | | 公開日 | 2011-03-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | X-ray crystal structure of transcriptional regulator (EF0465) from Enterococcus faecalis, Northeast Structural Genomics Consortium Target EfR190
To be Published
|
|
3W94
 
 | |
2RA2
 
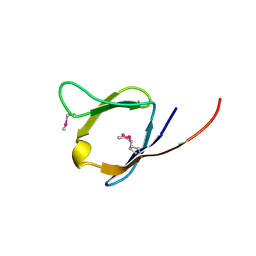 | | X-Ray structure of the Q7CPV8 protein from Salmonella typhimurium at the resolution 1.9 A. Northeast Structural Genomics Consortium target StR88A | | 分子名称: | Putative lipoprotein | | 著者 | Kuzin, A.P, Su, M, Seetharaman, J, Vorobiev, S.M, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-09-14 | | 公開日 | 2007-10-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-Ray structure of the Q7CPV8 protein from Salmonella typhimurium at the resolution 1.9 A.
To be Published
|
|
2RAD
 
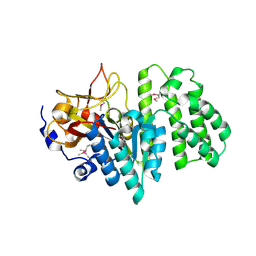 | | Crystal structure of the succinoglycan biosynthesis protein. Northeast structural Genomics Consortium target BcR135 | | 分子名称: | Succinoglycan biosynthesis protein | | 著者 | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-09-14 | | 公開日 | 2007-10-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of the succinoglycan biosynthesis protein.
To be Published
|
|
2RB6
 
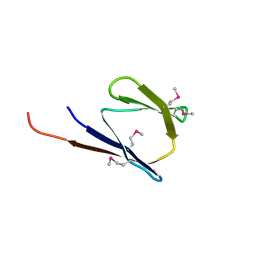 | | X-Ray structure of the protein Q8EI81. Northeast Structural Genomics Consortium target SoR78A | | 分子名称: | Uncharacterized protein | | 著者 | Kuzin, A.P, Su, M, Seetharaman, J, Vorobiev, S.M, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-09-18 | | 公開日 | 2007-10-23 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-Ray structure of the protein Q8EI81.
To be Published
|
|
3QWE
 
 | | Crystal structure of the N-terminal domain of the GEM interacting protein | | 分子名称: | GEM-interacting protein, UNKNOWN ATOM OR ION | | 著者 | Guan, X, Tempel, W, Tong, Y, Shen, L, Wang, H, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2011-02-28 | | 公開日 | 2011-03-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of the GEM interacting protein
to be published
|
|
4KUL
 
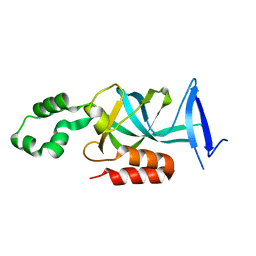 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | 分子名称: | Regulatory protein SIR3 | | 著者 | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | 登録日 | 2013-05-22 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUI
 
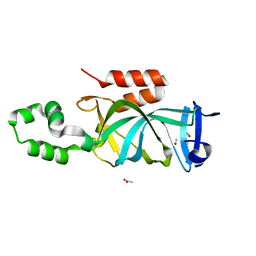 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | 分子名称: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | 著者 | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | 登録日 | 2013-05-22 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | 分子名称: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | 著者 | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | 登録日 | 2013-05-22 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.203 Å) | | 主引用文献 | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2WOC
 
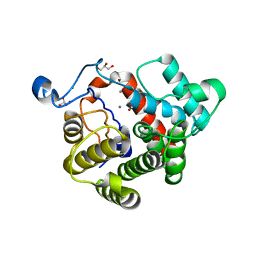 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum | | 分子名称: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | 登録日 | 2009-07-23 | | 公開日 | 2009-08-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOE
 
 | | Crystal Structure of the D97N variant of dinitrogenase reductase- activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP-ribose | | 分子名称: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | 登録日 | 2009-07-23 | | 公開日 | 2009-08-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOD
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP- ribsoyllysine | | 分子名称: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | 登録日 | 2009-07-23 | | 公開日 | 2009-08-11 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
