6DLU
 
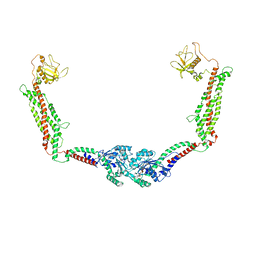 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
6DLV
 
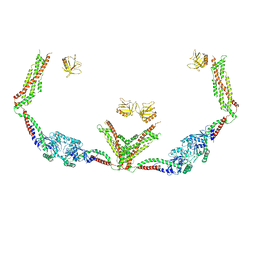 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
7T2G
 
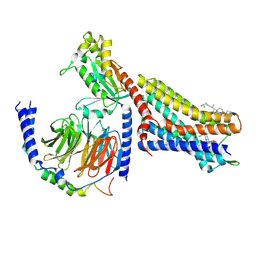 | | CryoEM structure of mu-opioid receptor - Gi protein complex bound to mitragynine pseudoindoxyl (MP) | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Seven, A.B, Qu, Q, Robertson, M.J, Wang, H, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2021-12-04 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Insights into distinct signaling profiles of the mu OR activated by diverse agonists.
Nat.Chem.Biol., 2022
|
|
7TN3
 
 | |
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | Authors: | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
3G3N
 
 | | PDE7A catalytic domain in complex with 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one | | Descriptor: | 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Castano, T, Wang, H. | | Deposit date: | 2009-02-02 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, structural analysis, and biological evaluation of thioxoquinazoline derivatives as phosphodiesterase 7 inhibitors
Chemmedchem, 4, 2009
|
|
5WH6
 
 | | Crystal structure of PDE4D2 in complex with inhibitor (S_Zl-n-91) | | Descriptor: | 1-[4-(difluoromethoxy)-3-{[(3S)-oxolan-3-yl]oxy}phenyl]-3-methylbutan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H, Wang, H. | | Deposit date: | 2017-07-14 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a PDE4-Specific Pocket for the Design of Selective Inhibitors.
Biochemistry, 57, 2018
|
|
3ISP
 
 | | Crystal structure of ArgP from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator Rv1985c/MT2039 | | Authors: | Zhou, X, Lou, Z, Sheng, F, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2009-08-27 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of ArgP from Mycobacterium tuberculosis Confirms Two Distinct Conformations of Full-length LysR Transcriptional Regulators and Reveals Its Function in DNA Binding and Transcriptional Regulation.
J.Mol.Biol., 2009
|
|
1A8O
 
 | | HIV CAPSID C-TERMINAL DOMAIN | | Descriptor: | HIV CAPSID | | Authors: | Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
1A43
 
 | | STRUCTURE OF THE HIV-1 CAPSID PROTEIN DIMERIZATION DOMAIN AT 2.6A RESOLUTION | | Descriptor: | HIV-1 CAPSID | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-02-10 | | Release date: | 1999-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BAJ
 
 | | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AUM
 
 | | HIV CAPSID C-TERMINAL DOMAIN (CAC146) | | Descriptor: | HIV CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I. | | Deposit date: | 1997-08-29 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
1BC9
 
 | | CYTOHESIN-1/B2-1 SEC7 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOHESIN-1 | | Authors: | Betz, S.F, Schnuchel, A, Wang, H, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1998-05-06 | | Release date: | 1999-05-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cytohesin-1 (B2-1) Sec7 domain and its interaction with the GTPase ADP ribosylation factor 1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8FU6
 
 | | GCGR-Gs complex in the presence of RAMP2 | | Descriptor: | Glucagon derivative ZP3780, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, O'Brien, E.S, Wang, H, Montabana, E, Kobilka, B.K. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Negative allosteric modulation of the glucagon receptor by RAMP2.
Cell, 186, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
1CKR
 
 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | HEAT SHOCK SUBSTRATE BINDING DOMAIN OF HSC-70 | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
4TMP
 
 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
4H58
 
 | | BRAF in complex with compound 3 | | Descriptor: | CHLORIDE ION, N-(4-{[(2-methoxyethyl)amino]methyl}phenyl)-6-(pyridin-4-yl)quinazolin-2-amine, Serine/threonine-protein kinase B-raf | | Authors: | Vasbinder, M, Aquila, B, Augustin, M, Chueng, T, Cook, D, Drew, L, Fauber, B, Glossop, S, Godin, R, Grondine, M, Hennessy, E, Johannes, J, Lee, S, Lyne, P, Moertl, M, Omer, C, Palakurthi, S, Pontz, T, Read, J, Sha, L, Shen, M, Steinbacher, S, Wang, H, Wu, A, Ye, M, Bagal, B. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf(V600E) Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
6KH4
 
 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
6KH3
 
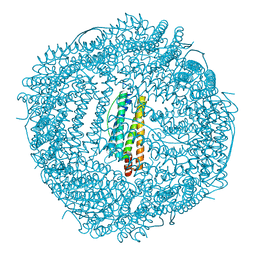 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
