7ZHI
 
 | |
7ZGS
 
 | |
7ZP2
 
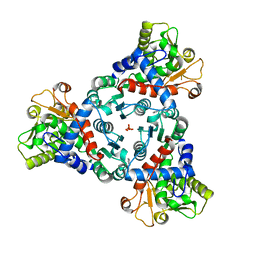
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with BDA-04 | | Descriptor: | Aspartate carbamoyltransferase, SODIUM ION, SULFATE ION, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7ZST
 
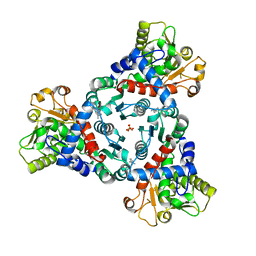
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with FLA-01 | | Descriptor: | 2-azanyl-~{N}-(2-methoxyethyl)-5-phenyl-thiophene-3-carboxamide, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-05-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7ZID
 
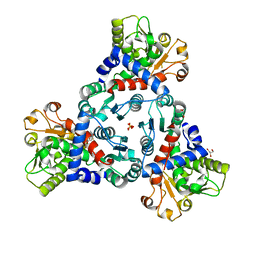 | |
8I02
 
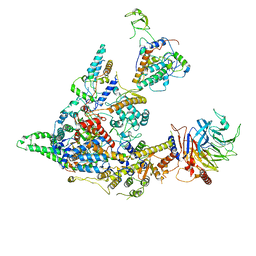 | | Cryo-EM structure of the SIN3S complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Histone deacetylase clr6, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
8I03
 
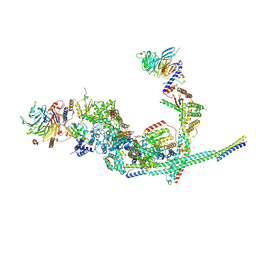 | | Cryo-EM structure of the SIN3L complex from S. pombe | | Descriptor: | Chromatin modification-related protein png2, Histone deacetylase clr6, POTASSIUM ION, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
7DL9
 
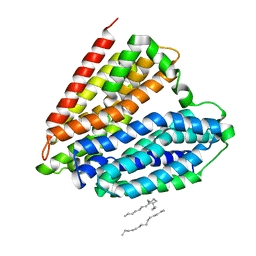 | | Crystal structure of nucleoside transporter NupG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Nucleoside permease NupG | | Authors: | Wang, C, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for substrate recognition by the bacterial nucleoside transporter NupG.
J.Biol.Chem., 296, 2021
|
|
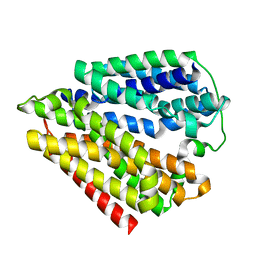
7DLA
 
 | |
9BRY
 
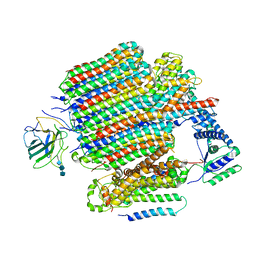 | | V0-only V-ATPase in synaptophysin gene knock-out mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRR
 
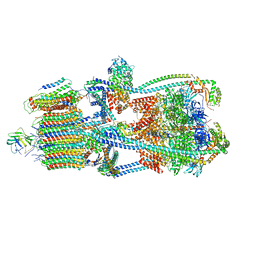 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRU
 
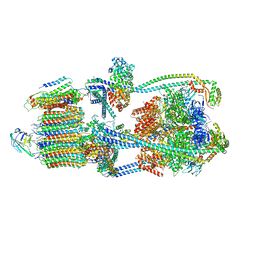 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRS
 
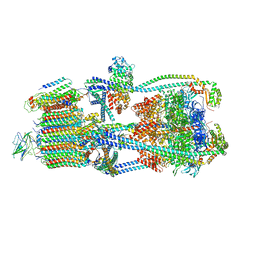 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRT
 
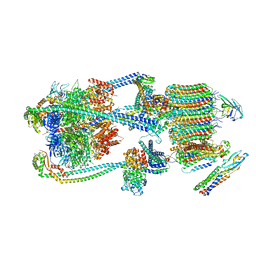 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRZ
 
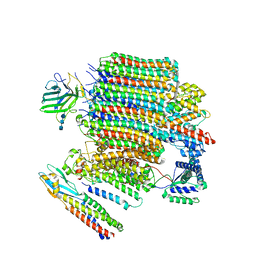 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
8W9D
 
 | |
8W9C
 
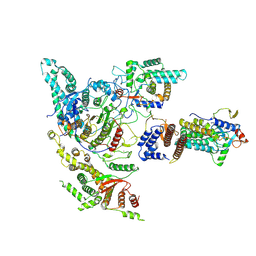 | |
8W9F
 
 | |
8W9E
 
 | |
1F0Z
 
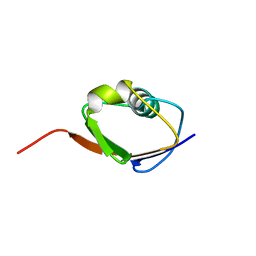 | | SOLUTION STRUCTURE OF THIS, THE SULFUR CARRIER PROTEIN IN E.COLI THIAMIN BIOSYNTHESIS | | Descriptor: | THIS PROTEIN | | Authors: | Wang, C, Xi, J, Begley, T.P, Nicholson, L.K. | | Deposit date: | 2000-05-17 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ThiS and implications for the evolutionary roots of ubiquitin.
Nat.Struct.Biol., 8, 2001
|
|
7D7N
 
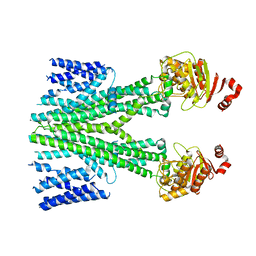 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
7D7R
 
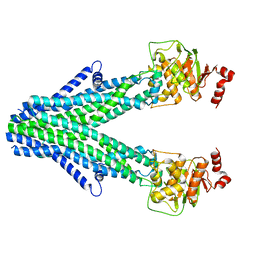 | | Cryo-EM structure of the core domain of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
5E8L
 
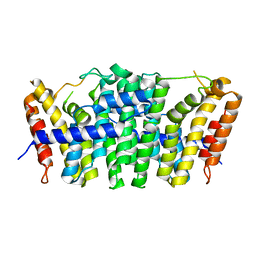 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | Descriptor: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
