3JCM
 
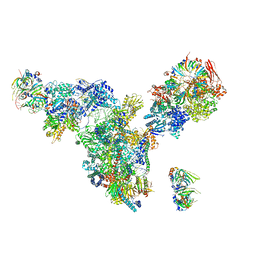 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
5GMK
 
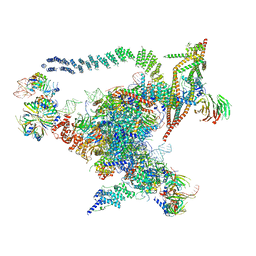 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | Descriptor: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
5Y88
 
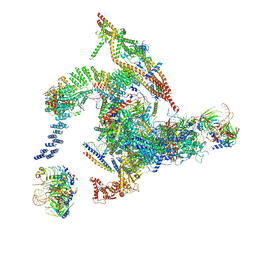 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-08-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
6J6G
 
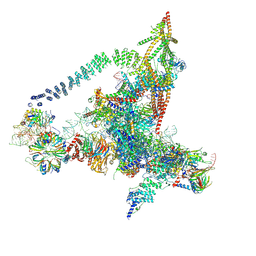 | | Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6Q
 
 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
6UM1
 
 | | Structure of M-6-P/IGFII Receptor at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, ... | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
8ZC9
 
 | | The Cryo-EM structure of DSR2-Tail tube-NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein, tail tube protein | | Authors: | Wang, R, Xu, Q, Wu, Z, Li, J, Yang, R, Shi, Z, Li, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
5J5C
 
 | | Crystal structure of ARL1-GTP and DCB domain of BIG1 complex | | Descriptor: | ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, R, Wang, Z, Zhang, T, Ding, J. | | Deposit date: | 2016-04-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for targeting BIG1 to Golgi apparatus through interaction of its DCB domain with Arl1
J Mol Cell Biol, 2016
|
|
5JHG
 
 | | Crystal structure of the complex between the human RhoA and the DH/PH domain of human ARHGEF11 | | Descriptor: | GLYCEROL, Rho guanine nucleotide exchange factor 11, Transforming protein RhoA | | Authors: | Wang, R, Chen, Q, Zhang, H, Yan, Z, Li, J, Miao, L, Wang, F. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA bound with its inhibitor and ARHGEF11
To Be Published
|
|
6XBY
 
 | | Cryo-EM structure of V-ATPase from bovine brain, state 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
6XBW
 
 | | Cryo-EM structure of V-ATPase from bovine brain, state 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
8ILB
 
 | | The complexes of RbcL, AtRaf1 and AtBSD2 (LFB) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8ILM
 
 | | The cryo-EM structure of eight Rubisco large subunits (RbcL), two Arabidopsis thaliana Rubisco accumulation factors 1 (AtRaf1), and seven Arabidopsis thaliana Bundle Sheath Defective 2 (AtBSD2) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IO2
 
 | | The Rubisco assembly intermidate of Arabidopsis thaliana Rubisco accumulation factor 1 (AtRaf1) and Rubisco large subunit (RbcL) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1.2, chloroplastic | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-10 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IOJ
 
 | | The Rubisco assembly intermidiate of Rubisco large subunit (RbcL) and Arabidopsis thaliana Rubisco accumulation factor 1 (AtRaf1) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1.2, chloroplastic | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IOL
 
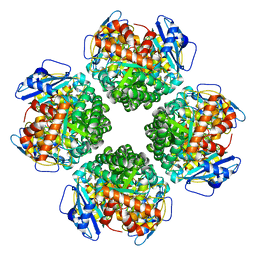 | | The complex of Rubisco large subunit (RbcL) | | Descriptor: | Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
6UM2
 
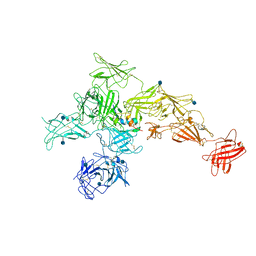 | | Structure of M-6-P/IGFII Receptor and IGFII complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, Insulin-like growth factor II | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
8BB7
 
 | |
7SHN
 
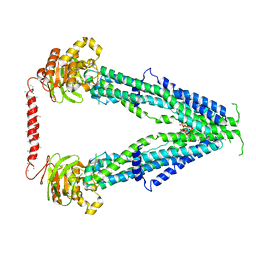 | | Cryo-EM structure of oleoyl-CoA-bound human peroxisomal fatty acid transporter ABCD1 | | Descriptor: | ATP-binding cassette sub-family D member 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Wang, R, Li, X. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of acyl-CoA transport across the peroxisomal membrane by human ABCD1.
Cell Res., 32, 2022
|
|
8B3K
 
 | |
7SHM
 
 | |
