5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1D1W
 
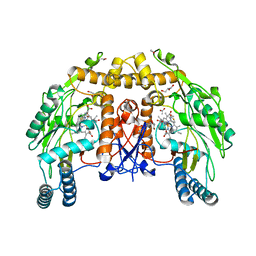 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 2-AMINOTHIAZOLINE (H4B BOUND) | | Descriptor: | 2-AMINOTHIAZOLINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Raman, C.S, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
5Y6D
 
 | |
5Y6E
 
 | |
6AE8
 
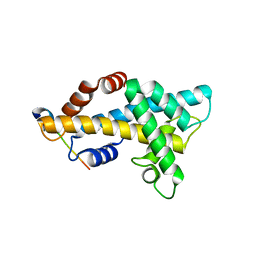 | | Structure insight into histone chaperone Chz1-mediated H2A.Z recognition and replacement | | Descriptor: | BICINE, Histone H2A.Z-specific chaperone CHZ1, Histone H2B.1,Histone H2A.Z | | Authors: | Wang, Y.Y, Shan, S, Zhou, Z. | | Deposit date: | 2018-08-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into histone chaperone Chz1-mediated H2A.Z recognition and histone replacement.
Plos Biol., 17, 2019
|
|
5GXJ
 
 | | Zika Virus NS2B-NS3 protease | | Descriptor: | FLAVIVIRUS_NS2B,LINKER,Peptidase S7 | | Authors: | Yang, H, Chen, X, Ji, X, Xiong, Y, Yang, K. | | Deposit date: | 2016-09-18 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Viral protease activation mechanism
To Be Published
|
|
4AGW
 
 | | Discovery of a small molecule type II inhibitor of wild-type and gatekeeper mutants of BCR-ABL, PDGFRalpha, Kit, and Src kinases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromet hyl)phenyl}benzamide, GLYCEROL, ... | | Authors: | Weisberg, E, Choi, H.G, Seeliger, M, Gray, N, Griffin, J.D. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-15 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Small-Molecule Type II Inhibitor of Wild-Type and Gatekeeper Mutants of Bcr-Abl, Pdgfralpha, Kit, and Src Kinases: Novel Type II Inhibitor of Gatekeeper Mutants.
Blood, 115, 2010
|
|
7D2S
 
 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
5XLT
 
 | | The crystal structure of tubulin in complex with 4'-demethylepipodophyllotoxin | | Descriptor: | (5S,5aR,8aR,9R)-9-(3,5-dimethoxy-4-oxidanyl-phenyl)-5-oxidanyl-5a,6,8a,9-tetrahydro-5H-[2]benzofuro[6,5-f][1,3]benzodioxol-8-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Structure of 4'-demethylepipodophyllotoxin in complex with tubulin provides a rationale for drug design
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5YQN
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor L55 | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[3-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQO
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor L5C | | Descriptor: | N-[4-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQM
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A29 | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-(4-phenylsulfanylphenyl)ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQL
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A2I | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[3-(phenoxymethyl)phenyl]ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8ITU
 
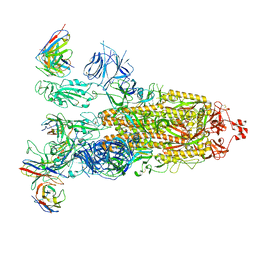 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 IgG. | | Descriptor: | 1H1 heavy chain, 1H1 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2023-03-23 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants
Commun Biol, 6, 2023
|
|
8IK3
 
 | |
8IK0
 
 | |
8J69
 
 | |
8NSE
 
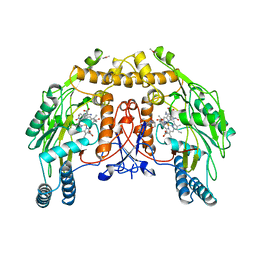 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, NNA COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-14 | | Release date: | 2001-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
3HDN
 
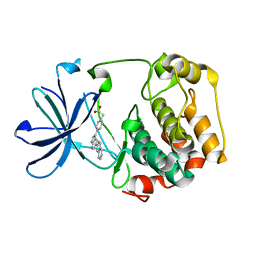 | |
3HDM
 
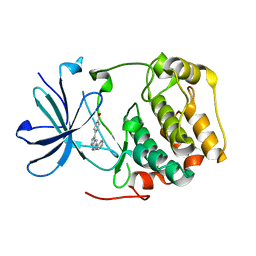 | |
8Q5I
 
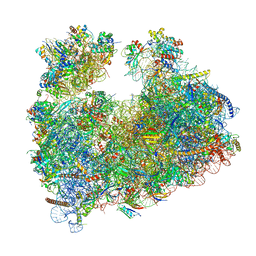 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | Descriptor: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
8P6Q
 
 | | Racemic structure of TNFR1 cysteine-rich domain | | Descriptor: | D-TNFR-1 CRD2, SULFATE ION, Tumor necrosis factor-binding protein 1 | | Authors: | Lander, A.J, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2023-05-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deciphering the Synthetic and Refolding Strategy of a Cysteine-Rich Domain in the Tumor Necrosis Factor Receptor (TNF-R) for Racemic Crystallography Analysis and d-Peptide Ligand Discovery.
Acs Bio Med Chem Au, 4, 2024
|
|
3KMW
 
 | | Crystal structure of the ILK/alpha-parvin core complex (MgATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-parvin, Integrin-linked kinase, ... | | Authors: | Fukuda, K, Qin, J. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions.
Mol.Cell, 36, 2009
|
|
