6VPM
 
 | |
6VPI
 
 | |
6VPL
 
 | |
6VMT
 
 | |
6VPH
 
 | |
6NZG
 
 | | Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine | | Descriptor: | (1S,2R,3S,4S,5S,6R)-2-amino-3,4,5,6-tetrahydroxycyclohexane-1-carboxylic acid, Beta-galactosidase, CALCIUM ION, ... | | Authors: | Pellock, S.J, Jariwala, P.B, Redinbo, M.R. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovering the Microbial Enzymes Driving Drug Toxicity with Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6VPJ
 
 | |
1NCC
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1NCD
 
 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
1NCA
 
 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
1NCB
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
3BCE
 
 | | Crystal structure of the ErbB4 kinase | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Receptor tyrosine-protein kinase erbB-4, ... | | Authors: | Qiu, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase.
Structure, 16, 2008
|
|
3BJE
 
 | |
3BBW
 
 | |
1NMB
 
 | | THE STRUCTURE OF A COMPLEX BETWEEN THE NC10 ANTIBODY AND INFLUENZA VIRUS NEURAMINIDASE AND COMPARISON WITH THE OVERLAPPING BINDING SITE OF THE NC41 ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FAB NC10, ... | | Authors: | Malby, R.L, Tulip, W.R, Colman, P.M. | | Deposit date: | 1995-01-17 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody
Structure, 2, 1994
|
|
3BBT
 
 | | crystal structure of the ErbB4 kinase in complex with lapatinib | | Descriptor: | N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Qiu, C. | | Deposit date: | 2007-11-11 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase.
Structure, 16, 2008
|
|
3C1Q
 
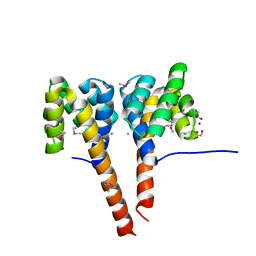 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Abendroth, J, Mitchell, D.D, Korotkov, K.V, Kreeger, A, Hol, W.G.J. | | Deposit date: | 2008-01-24 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae
J.Struct.Biol., 166, 2009
|
|
4M97
 
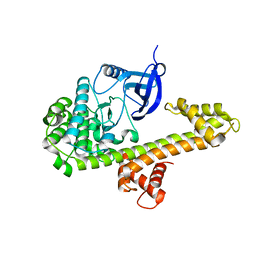 | | Calcium-Dependent Protein Kinase 1 from Neospora caninum | | Descriptor: | Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-08-14 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
3C57
 
 | |
3BZ3
 
 | |
3C3W
 
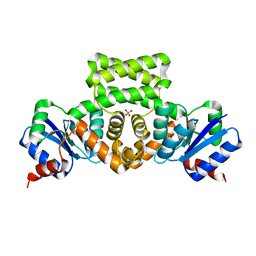 | | Crystal Structure of the Mycobacterium tuberculosis Hypoxic Response Regulator DosR | | Descriptor: | SULFATE ION, TWO COMPONENT TRANSCRIPTIONAL REGULATORY PROTEIN DEVR | | Authors: | Wisedchaisri, G, Wu, M, Sherman, D.R, Hol, W.G.J. | | Deposit date: | 2008-01-28 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation
J.Mol.Biol., 378, 2008
|
|
4MXA
 
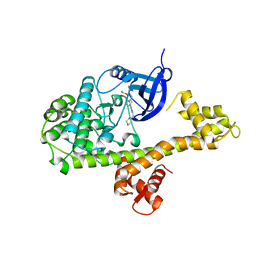 | | CDPK1 from Neospora caninum in complex with inhibitor RM-1-132 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
3CVL
 
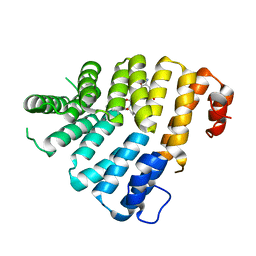 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphofructokinase (PFK) PTS1 peptide | | Descriptor: | Peroxisome targeting signal 1 receptor PEX5, T. brucei PFK PTS1 peptide Ac-HEELAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVQ
 
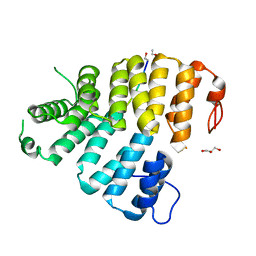 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
1EFI
 
 | |
