2MCV
 
 | | Solid-state NMR structure of piscidin 1 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Hayden, R.M, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
5J30
 
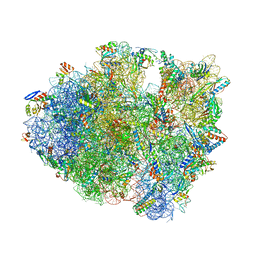 | |
5J3C
 
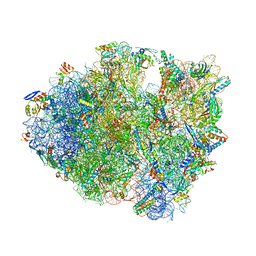 | |
2MCU
 
 | | Solid-state NMR structure of piscidin 1 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2HIO
 
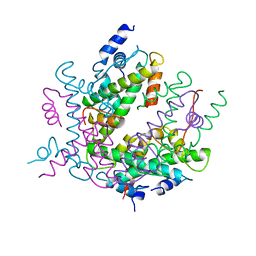 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN | | Descriptor: | PROTEIN (HISTONE H2A), PROTEIN (HISTONE H2B), PROTEIN (HISTONE H3), ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1999-06-15 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1I2Z
 
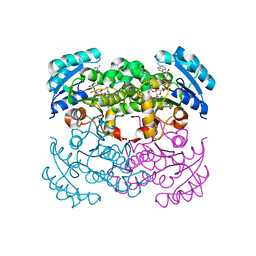 | | E. COLI ENOYL REDUCTASE IN COMPLEX WITH NAD AND BRL-12654 | | Descriptor: | 4-(2-THIENYL)-1-(4-METHYLBENZYL)-1H-IMIDAZOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Heerding, D.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl acyl carrier protein
reductase (FabI).
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1I30
 
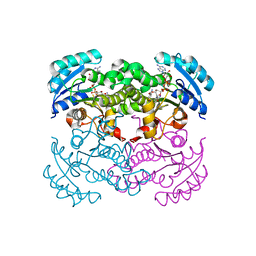 | | E. Coli Enoyl Reductase +NAD+SB385826 | | Descriptor: | 1,3,4,9-TETRAHYDRO-2-(HYDROXYBENZOYL)-9-[(4-HYDROXYPHENYL)METHYL]-6-METHOXY-2H-PYRIDO[3,4-B]INDOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Seefeld, M.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial enoyl acyl carrier protein reductase (FabI): 2,9-disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
3PSU
 
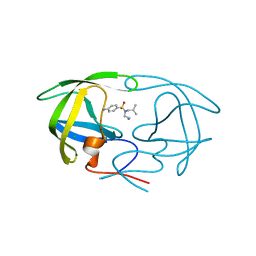 | |
3PSY
 
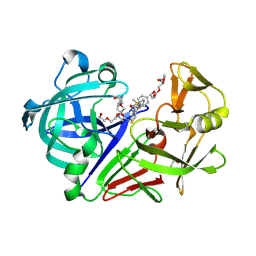 | | Endothiapepsin in complex with an inhibitor based on the Gewald reaction | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3QBF
 
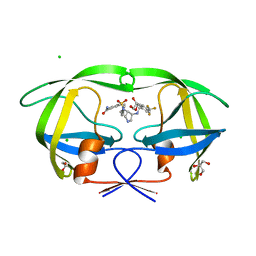 | |
3QIH
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis(3-hydroxybenzyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease, ... | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-01-27 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Novel inhibitors for HIV-1 protease
To be Published
|
|
3QN8
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis(3-hydroxybenzyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Novel inhibitors for HIV-1 protease
To be Published
|
|
3QOB
 
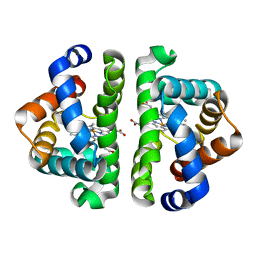 | |
3QRM
 
 | |
3QP0
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis[3-(hydroxymethyl)benzyl]hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-02-11 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel inhibitors for HIV-1 protease
To be Published
|
|
3QRO
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a three-armed pyrrolidine-based inhibitor | | Descriptor: | 4-({(3S,4S)-4-[(3,5-dihydroxybenzyl)amino]pyrrolidin-3-yl}[4-(trifluoromethyl)benzyl]sulfamoyl)benzamide, CHLORIDE ION, DITHIANE DIOL, ... | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.616 Å) | | Cite: | Design of a series of novel three-armed pyrrolidine-based inhibitors for HIV-1 protease
To be Published
|
|
3QPJ
 
 | |
3QRS
 
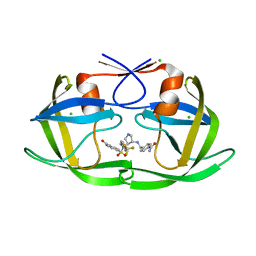 | |
3T6I
 
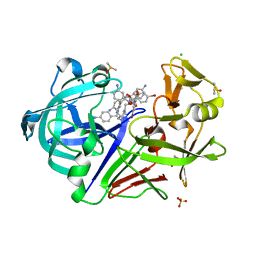 | | Endothiapepsin in complex with an azepin derivative | | Descriptor: | (3R)-3-({(4-aminobenzyl)[(4-aminophenyl)acetyl]amino}methyl)-5-(hydroxymethyl)-2,3,4,7-tetrahydro-1H-azepinium, (3R)-3-({(4-aminobenzyl)[(4-aminophenyl)acetyl]amino}methyl)-5-{[(4-bromobenzoyl)oxy]methyl}-2,3,4,7-tetrahydro-1H-azepinium, 4-bromobenzoic acid, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Endothiapepsin in complex with an azepin derivative
To be Published
|
|
5T4Z
 
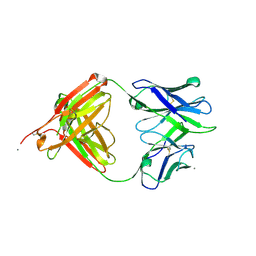 | |
5U0U
 
 | |
5U7L
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | (3R)-1-{3-[5-(4-ethylphenyl)-1-methyl-1H-pyrazol-4-yl]-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl}-N,N-dimethylpyrrolidin-3-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
5U7I
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 4-[3-(4-methoxyphenoxy)azetidin-1-yl]-1-methyl-3-(2-methylpropyl)-1H-pyrazolo[3,4-d]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
5U15
 
 | |
5TRP
 
 | |
